transcription factor AP-2 delta (activating enhancer binding protein 2 delta)
ZFIN 
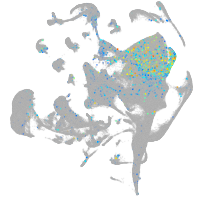
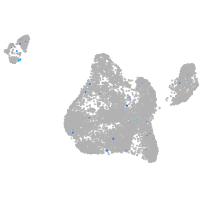
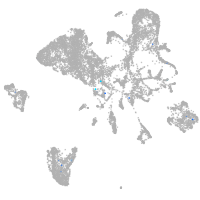
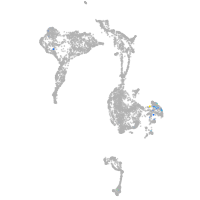
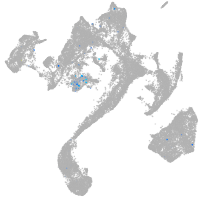
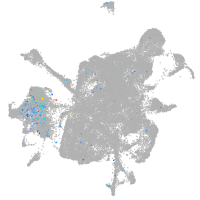
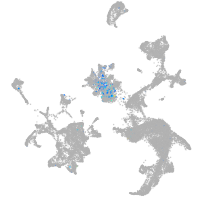
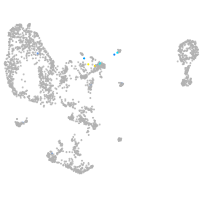
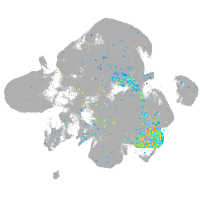
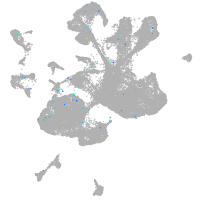
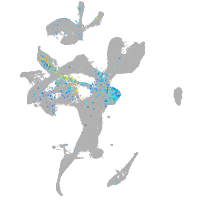
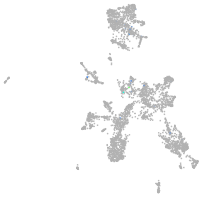
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| celsr3 | 0.354 | aldh9a1a.1 | -0.028 |
| si:dkeyp-69c1.7 | 0.341 | pgk1 | -0.028 |
| CR388190.1 | 0.312 | tob1a | -0.027 |
| LOC797104 | 0.304 | pgam1a | -0.027 |
| pou4f3 | 0.263 | rgrb | -0.026 |
| tmem151bb | 0.254 | glud1b | -0.026 |
| atoh7 | 0.227 | tuba8l2 | -0.026 |
| LOC101882645 | 0.205 | hadh | -0.025 |
| susd1 | 0.198 | eno3 | -0.025 |
| XLOC-031363 | 0.194 | scp2a | -0.025 |
| hes2.1 | 0.194 | atp5mc1 | -0.025 |
| XLOC-018000 | 0.190 | pgm1 | -0.025 |
| zgc:110045 | 0.183 | pgrmc1 | -0.024 |
| zgc:158291 | 0.183 | mgst1.2 | -0.024 |
| hecw2a | 0.181 | hspd1 | -0.024 |
| mkrn2os.2 | 0.177 | CABZ01032488.1 | -0.024 |
| LOC100535857 | 0.175 | ndufa7 | -0.024 |
| pou4f2 | 0.171 | gpia | -0.024 |
| barhl2 | 0.170 | ndufv1 | -0.024 |
| gpsm1a | 0.160 | mdh1aa | -0.024 |
| mid1 | 0.152 | gcsha | -0.023 |
| inafm2 | 0.152 | ndufa12 | -0.023 |
| si:ch211-153f2.7 | 0.148 | akt2l | -0.023 |
| LOC110440109 | 0.146 | prdx6 | -0.023 |
| kif26bb | 0.145 | ostc | -0.023 |
| dhx32b | 0.142 | ndufb8 | -0.023 |
| INSM2 | 0.142 | ndufs2 | -0.023 |
| LOC110437839 | 0.135 | nipsnap3a | -0.023 |
| nlgn3a | 0.135 | aldh6a1 | -0.023 |
| CABZ01076968.1 | 0.131 | alas1 | -0.023 |
| emb | 0.130 | COX7A2 (1 of many) | -0.023 |
| ebf3a | 0.127 | pcbd1 | -0.022 |
| LOC110439576 | 0.126 | g6pca.2 | -0.022 |
| brinp3a.1 | 0.126 | tecrb | -0.022 |
| XLOC-034192 | 0.125 | glod5 | -0.022 |