transcription factor AP-2 delta (activating enhancer binding protein 2 delta)
ZFIN 
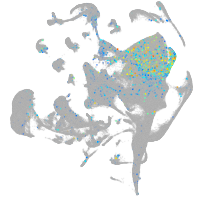
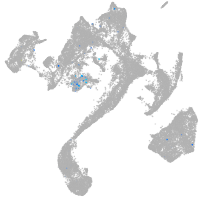
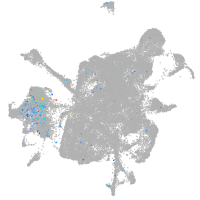
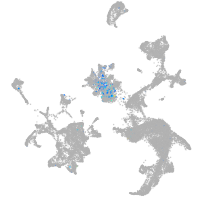
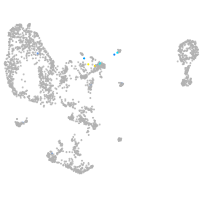
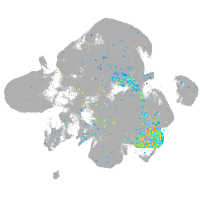
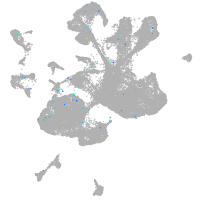
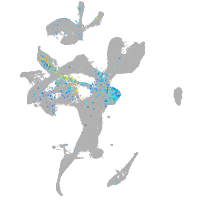
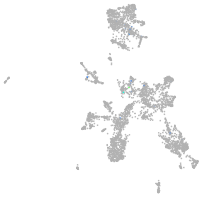
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| insm1a | 0.175 | pmp22a | -0.044 |
| stmn1b | 0.174 | si:dkey-151g10.6 | -0.035 |
| elavl3 | 0.171 | rps2 | -0.034 |
| rtn1a | 0.170 | rps24 | -0.034 |
| barhl2 | 0.170 | krt18a.1 | -0.033 |
| tuba1c | 0.169 | mfap2 | -0.032 |
| pou4f2 | 0.169 | rplp2l | -0.031 |
| gpm6aa | 0.161 | rps19 | -0.031 |
| lhx9 | 0.159 | rpl9 | -0.030 |
| pou4f1 | 0.158 | krt8 | -0.030 |
| lmo3 | 0.151 | id3 | -0.029 |
| islr2 | 0.151 | rplp1 | -0.029 |
| fez1 | 0.146 | fkbp14 | -0.029 |
| tuba1a | 0.146 | rpl18 | -0.029 |
| tfap2e | 0.144 | tpm4a | -0.029 |
| kif3cb | 0.143 | rps13 | -0.028 |
| rtn1b | 0.141 | rps8a | -0.027 |
| mllt11 | 0.141 | sdc4 | -0.027 |
| scrt2 | 0.141 | rps27a | -0.027 |
| gpm6ab | 0.141 | sparc | -0.027 |
| XLOC-018000 | 0.140 | si:ch211-286o17.1 | -0.026 |
| tmsb | 0.138 | crtap | -0.026 |
| ppp1r14ba | 0.135 | id1 | -0.026 |
| thsd7aa | 0.135 | rack1 | -0.025 |
| myt1a | 0.134 | ahnak | -0.025 |
| olfm1b | 0.134 | rpl26 | -0.025 |
| nova2 | 0.133 | rps10 | -0.025 |
| gng3 | 0.133 | twist1a | -0.025 |
| zgc:153426 | 0.129 | rpl13a | -0.025 |
| pou3f1 | 0.128 | rpl3 | -0.025 |
| si:ch211-137a8.4 | 0.123 | eef1da | -0.025 |
| gap43 | 0.123 | rpl36 | -0.025 |
| tubb5 | 0.122 | p4ha1b | -0.024 |
| pou3f2b | 0.121 | rpl23 | -0.024 |
| sncb | 0.121 | rpl11 | -0.024 |