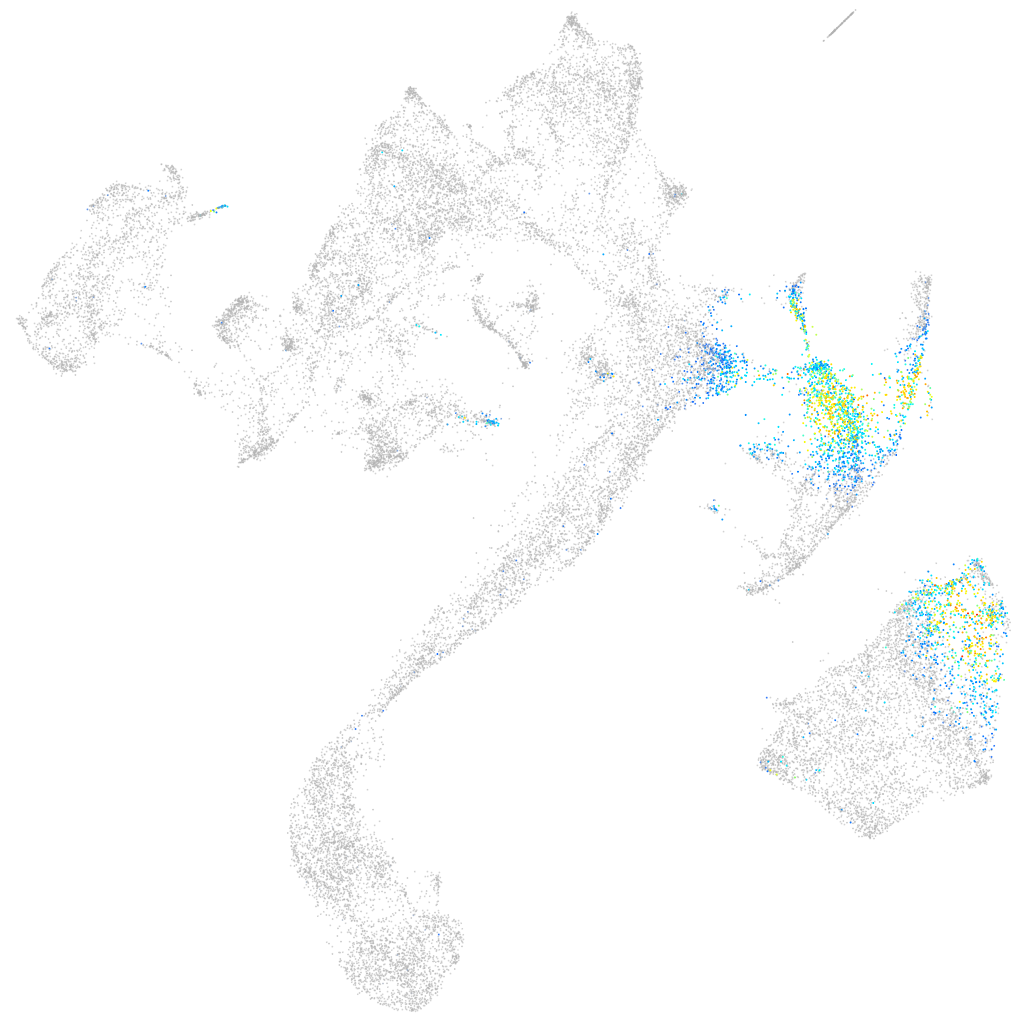
T-box transcription factor 6
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| ripply2 | 0.584 | fabp3 | -0.295 |
| pcdh8 | 0.561 | sparc | -0.226 |
| her11 | 0.515 | actc1b | -0.218 |
| myf5 | 0.498 | bhmt | -0.212 |
| mespab | 0.487 | pabpc4 | -0.205 |
| draxin | 0.474 | ckma | -0.197 |
| mespaa | 0.473 | atp2a1 | -0.197 |
| her1 | 0.456 | ak1 | -0.197 |
| gnaia | 0.441 | ckmb | -0.197 |
| grin2da | 0.434 | vwde | -0.197 |
| zic3 | 0.423 | eno1a | -0.196 |
| arl4aa | 0.421 | eef1da | -0.196 |
| apoc1 | 0.419 | col1a2 | -0.191 |
| efnb2b | 0.417 | tmem38a | -0.191 |
| nid2a | 0.416 | mylpfa | -0.190 |
| ripply1 | 0.392 | eif4a1b | -0.184 |
| fn1b | 0.388 | gamt | -0.183 |
| ism1 | 0.374 | pmp22a | -0.182 |
| fhdc2 | 0.369 | gapdh | -0.182 |
| dlc | 0.368 | col1a1a | -0.178 |
| meox1 | 0.367 | si:dkey-16p21.8 | -0.175 |
| igsf9a | 0.364 | tpi1b | -0.175 |
| sfrp1a | 0.357 | aldoab | -0.175 |
| foxc1a | 0.356 | myl1 | -0.174 |
| tuba8l2 | 0.352 | srl | -0.173 |
| BX001014.2 | 0.351 | pvalb1 | -0.172 |
| msgn1 | 0.349 | pvalb2 | -0.170 |
| her7 | 0.345 | ndrg2 | -0.170 |
| pstpip2 | 0.343 | acta1b | -0.170 |
| zfand5a | 0.342 | celf2 | -0.169 |
| large2 | 0.341 | neb | -0.169 |
| foxc1b | 0.331 | mylz3 | -0.169 |
| asph | 0.331 | FQ323156.1 | -0.168 |
| rgs12a | 0.325 | ldb3b | -0.168 |
| adgrl2a | 0.323 | CABZ01078594.1 | -0.167 |