Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line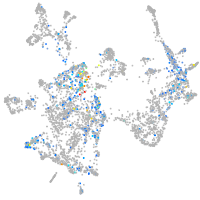 |
epidermis |
periderm |
fin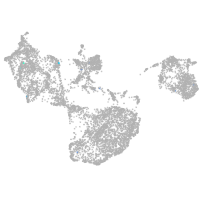 |
ionocytes / mucous |
pigment cells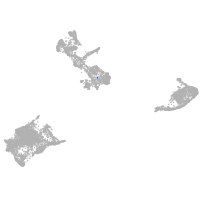 |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| scospondin | 0.426 | tmsb4x | -0.092 |
| ctgfa | 0.367 | ptmaa | -0.085 |
| slit1b | 0.339 | fabp3 | -0.083 |
| arxa | 0.335 | ptmab | -0.076 |
| aplnr2 | 0.334 | elavl3 | -0.074 |
| shhb | 0.327 | hmgb3a | -0.069 |
| col9a3 | 0.327 | hmgb1b | -0.069 |
| optc | 0.314 | hnrnpaba | -0.066 |
| col9a2 | 0.314 | rtn1a | -0.062 |
| myl9a | 0.310 | sox3 | -0.062 |
| shha | 0.305 | tmsb | -0.061 |
| clu | 0.299 | ckbb | -0.061 |
| cyr61l2 | 0.289 | si:ch211-137a8.4 | -0.057 |
| npr3 | 0.288 | marcksb | -0.056 |
| col8a1a | 0.287 | lfng | -0.056 |
| spon1b | 0.276 | CU467822.1 | -0.056 |
| sulf1 | 0.268 | myt1b | -0.055 |
| col9a1b | 0.267 | si:dkey-276j7.1 | -0.055 |
| fxyd1 | 0.266 | tubb2b | -0.055 |
| emilin3a | 0.264 | tmeff1b | -0.054 |
| b3gnt7l | 0.258 | sox11b | -0.054 |
| vasna | 0.257 | si:ch73-21g5.7 | -0.052 |
| trabd2b | 0.255 | si:dkey-56m19.5 | -0.051 |
| scinlb | 0.253 | prdx2 | -0.050 |
| anxa4 | 0.251 | si:ch211-288g17.3 | -0.050 |
| spon2a | 0.244 | mex3b | -0.050 |
| lcn15 | 0.240 | cadm3 | -0.050 |
| tbata | 0.238 | nova2 | -0.049 |
| slit2 | 0.236 | stmn1b | -0.049 |
| oc90 | 0.235 | mdkb | -0.048 |
| r3hdml | 0.235 | hnrnpa0b | -0.048 |
| foxa2 | 0.232 | pou3f3b | -0.048 |
| b4galnt3b | 0.231 | sb:cb81 | -0.048 |
| zgc:162730 | 0.231 | gas1b | -0.048 |
| wnt4b | 0.230 | hmgb1a | -0.048 |




