shadow of prion protein 2
ZFIN 
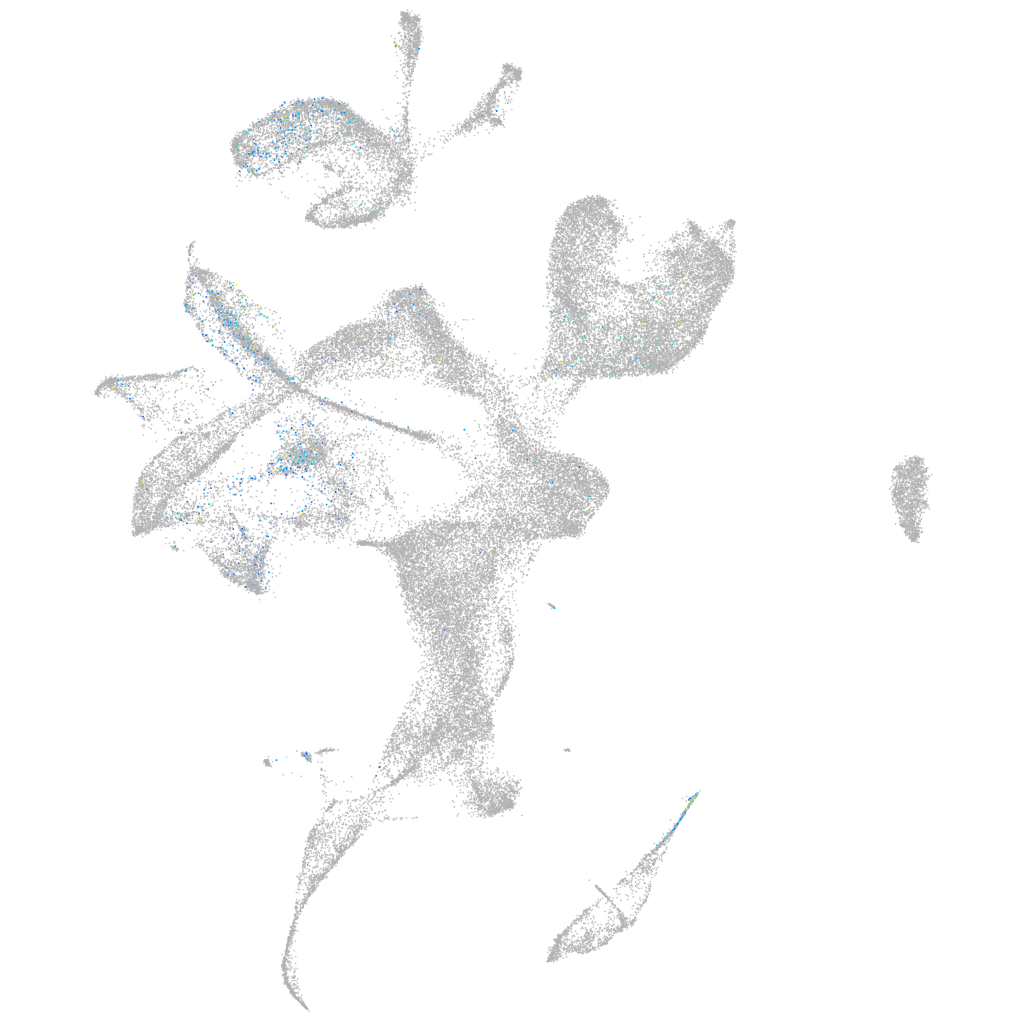
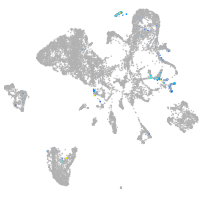
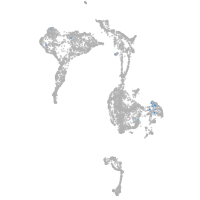
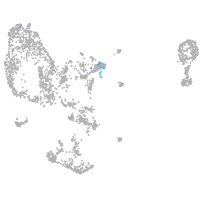
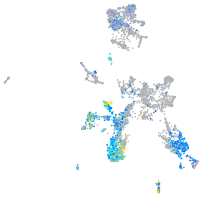
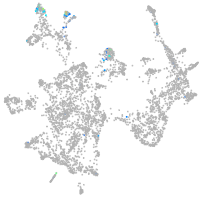
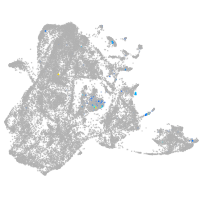
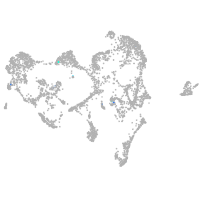
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| crygm2d1 | 0.236 | hmgb2a | -0.100 |
| crygm2d2 | 0.229 | slc25a5 | -0.064 |
| crygm2d18 | 0.228 | hmga1a | -0.063 |
| capn3a | 0.224 | pcna | -0.060 |
| crygm2d5 | 0.220 | hmgb2b | -0.060 |
| crygm2d3 | 0.220 | rrm1 | -0.059 |
| crygm2d4 | 0.218 | fabp7a | -0.059 |
| crygm2d20 | 0.217 | ahcy | -0.059 |
| crygm2d10 | 0.217 | mki67 | -0.055 |
| crygm2d7 | 0.217 | stmn1a | -0.053 |
| crygm2d16 | 0.215 | nutf2l | -0.053 |
| crygm2d11 | 0.214 | tuba8l | -0.053 |
| BX545855.1 | 0.214 | ptmab | -0.053 |
| crygm2d21 | 0.211 | ccnd1 | -0.052 |
| crygm2d8 | 0.210 | dut | -0.052 |
| crygm2d12 | 0.210 | chaf1a | -0.052 |
| crygm2d6 | 0.207 | si:ch73-1a9.3 | -0.052 |
| crygm2d13 | 0.206 | dek | -0.051 |
| crygm2d9 | 0.203 | seta | -0.051 |
| lctla | 0.199 | msi1 | -0.051 |
| crygm2d15 | 0.196 | ccna2 | -0.051 |
| lctlb | 0.192 | mdka | -0.050 |
| crygmxl2 | 0.185 | snrpd1 | -0.049 |
| crygm2d14 | 0.184 | rpa3 | -0.049 |
| endou2 | 0.180 | rrm2 | -0.049 |
| si:ch211-255g12.6 | 0.178 | lig1 | -0.049 |
| bfsp2 | 0.174 | hmgn2 | -0.049 |
| gpx9 | 0.168 | si:ch73-281n10.2 | -0.049 |
| lim2.5 | 0.166 | fen1 | -0.049 |
| crybb1l2 | 0.160 | her15.1 | -0.048 |
| cryaa | 0.160 | rpa2 | -0.048 |
| crybb1l1 | 0.159 | mcm7 | -0.048 |
| BFSP1 | 0.155 | selenoh | -0.048 |
| lim2.4 | 0.151 | cks1b | -0.047 |
| cryba1l2 | 0.149 | cx43.4 | -0.047 |