spen family transcriptional repressor
ZFIN 
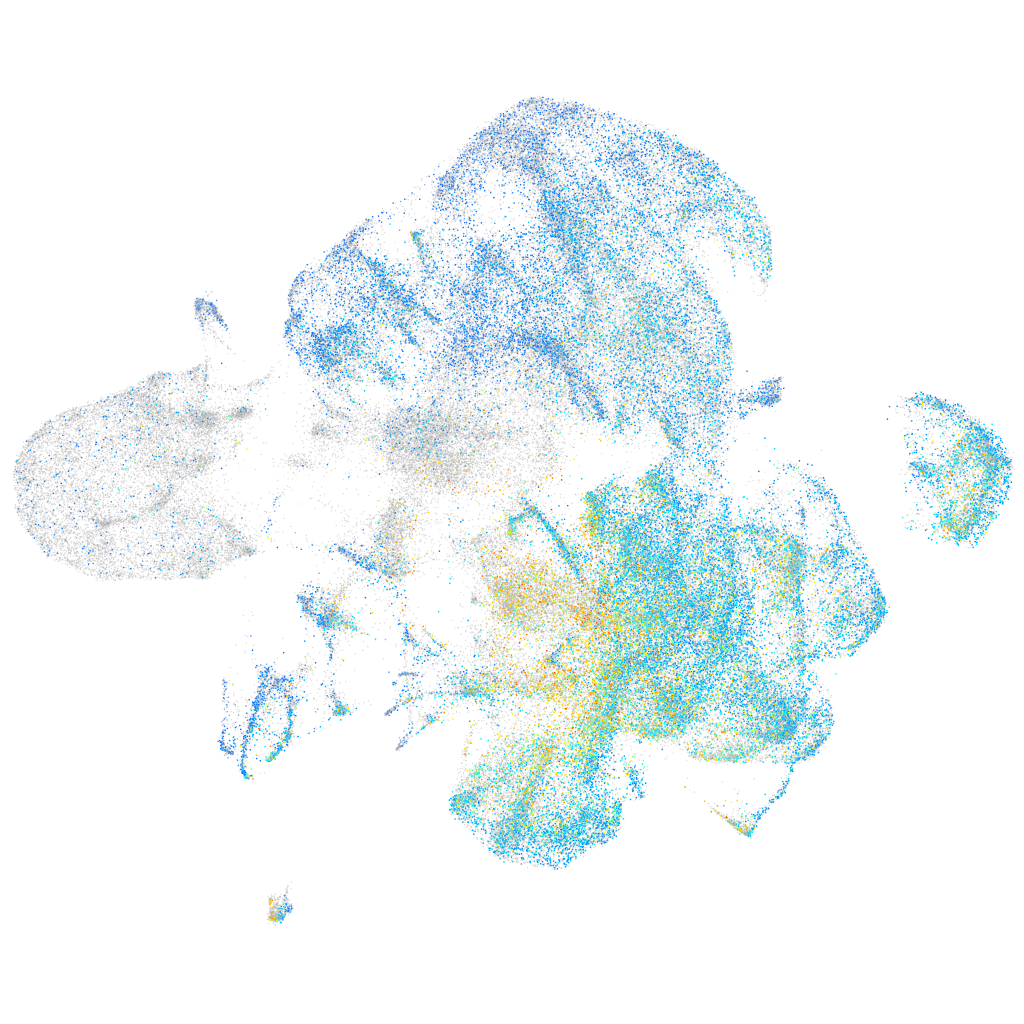
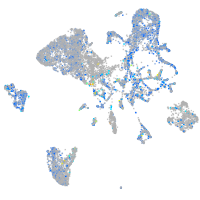
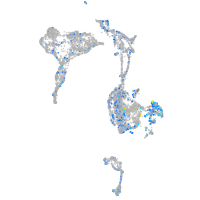
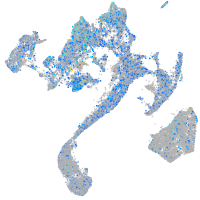
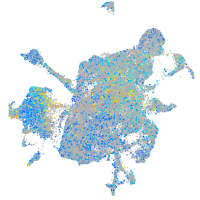
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| elavl4 | 0.244 | hmgb2a | -0.246 |
| stmn1b | 0.240 | hmga1a | -0.204 |
| sncb | 0.238 | npm1a | -0.196 |
| stx1b | 0.238 | pcna | -0.195 |
| gpm6aa | 0.234 | eef1a1l1 | -0.190 |
| snap25a | 0.233 | hmgb2b | -0.189 |
| gpm6ab | 0.233 | nop58 | -0.187 |
| tuba1c | 0.229 | si:dkey-151g10.6 | -0.186 |
| rtn1a | 0.227 | snu13b | -0.181 |
| gng3 | 0.226 | dkc1 | -0.179 |
| stmn2a | 0.224 | anp32b | -0.174 |
| vamp2 | 0.224 | fbl | -0.173 |
| stxbp1a | 0.223 | ranbp1 | -0.169 |
| ywhag2 | 0.222 | hspb1 | -0.168 |
| calm1a | 0.219 | banf1 | -0.168 |
| atp6v0cb | 0.216 | stmn1a | -0.167 |
| rtn1b | 0.215 | si:ch73-281n10.2 | -0.167 |
| ywhah | 0.215 | nop56 | -0.166 |
| CR383676.1 | 0.214 | rps12 | -0.166 |
| kdm6bb | 0.214 | rplp2l | -0.166 |
| elavl3 | 0.212 | id1 | -0.166 |
| ptmaa | 0.212 | chaf1a | -0.164 |
| ckbb | 0.205 | tuba8l4 | -0.163 |
| cdk5r1b | 0.203 | rsl1d1 | -0.163 |
| zgc:65894 | 0.200 | rps28 | -0.162 |
| gapdhs | 0.198 | nop10 | -0.161 |
| gnao1a | 0.198 | rplp1 | -0.161 |
| map1aa | 0.197 | ran | -0.160 |
| atp6v1e1b | 0.197 | lig1 | -0.160 |
| atp6v1g1 | 0.196 | mcm6 | -0.159 |
| tmsb2 | 0.195 | nhp2 | -0.158 |
| nova2 | 0.192 | selenoh | -0.158 |
| nrxn1a | 0.192 | snrpb | -0.158 |
| rnasekb | 0.191 | nasp | -0.154 |
| calm2a | 0.191 | snrpf | -0.152 |