spen family transcriptional repressor
ZFIN 
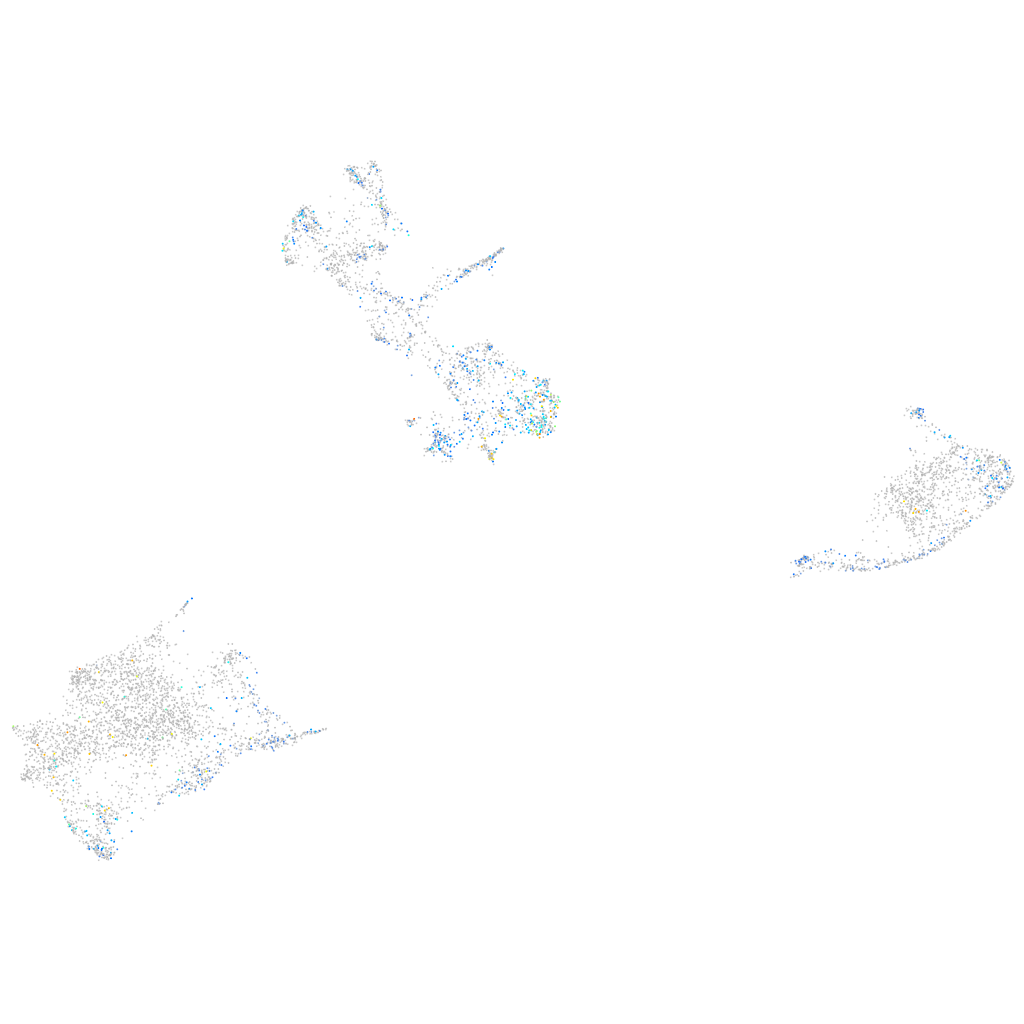
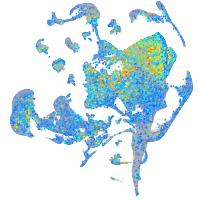
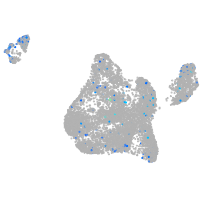
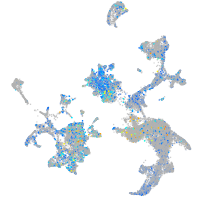
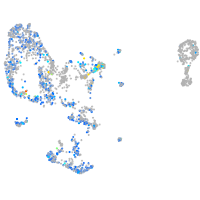
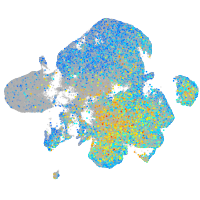
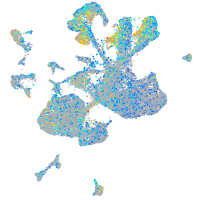
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| elavl3 | 0.210 | gstp1 | -0.119 |
| stx1b | 0.199 | gch2 | -0.102 |
| nova2 | 0.199 | pts | -0.096 |
| gpm6aa | 0.192 | actb2 | -0.095 |
| stmn1b | 0.188 | paics | -0.094 |
| rnasekb | 0.183 | akr1b1 | -0.093 |
| rtn1a | 0.182 | si:dkey-151g10.6 | -0.091 |
| vamp2 | 0.181 | zgc:110339 | -0.088 |
| pbx1a | 0.176 | prdx1 | -0.085 |
| camk2d2 | 0.172 | pfn1 | -0.083 |
| napgb | 0.169 | si:dkey-251i10.2 | -0.083 |
| atp6v0cb | 0.169 | gpd1b | -0.079 |
| fez1 | 0.169 | uraha | -0.077 |
| pbx3b | 0.168 | pcbd1 | -0.074 |
| elavl4 | 0.168 | gpr143 | -0.073 |
| zgc:65894 | 0.166 | slc22a7a | -0.072 |
| rbfox1 | 0.165 | cyb5a | -0.071 |
| sncb | 0.165 | CABZ01032488.1 | -0.070 |
| marcksl1a | 0.164 | qdpra | -0.069 |
| gng2 | 0.164 | impdh1b | -0.067 |
| sypb | 0.163 | rplp0 | -0.067 |
| gng3 | 0.162 | prdx5 | -0.067 |
| zfhx4 | 0.161 | CABZ01021592.1 | -0.065 |
| dscaml1 | 0.161 | rpl36a | -0.061 |
| CR383676.1 | 0.160 | rbp4l | -0.061 |
| nrxn1a | 0.160 | sult1st1 | -0.061 |
| PRKAR2B | 0.160 | tspan36 | -0.061 |
| sv2a | 0.160 | phyhd1 | -0.056 |
| ptmaa | 0.160 | mdh1aa | -0.055 |
| tmeff1b | 0.160 | C12orf75 | -0.053 |
| eno2 | 0.159 | prdx6 | -0.052 |
| epb41a | 0.159 | atic | -0.052 |
| snap25b | 0.159 | gstt1a | -0.051 |
| nsg2 | 0.159 | oacyl | -0.051 |
| stxbp1a | 0.159 | krt4 | -0.051 |