solute carrier family 4 member 4a
ZFIN 
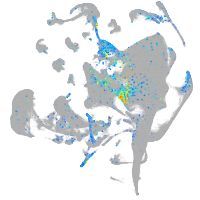
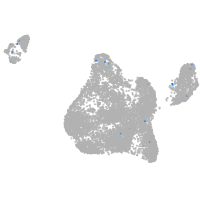
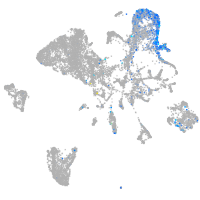
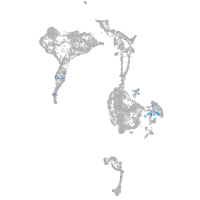
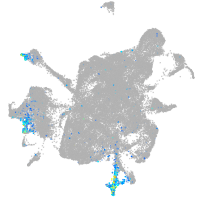
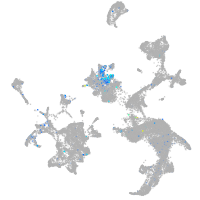
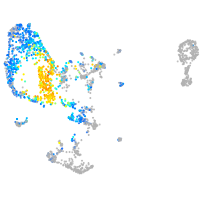
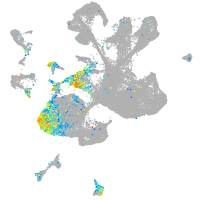
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| zpld1b | 0.264 | ahnak | -0.080 |
| zpld1a | 0.259 | zgc:153867 | -0.076 |
| otog | 0.254 | krt8 | -0.074 |
| chl1b | 0.226 | tfap2a | -0.073 |
| FO203514.1 | 0.220 | mcama | -0.072 |
| hhla1 | 0.213 | meis1a | -0.072 |
| FAM163A | 0.197 | gsnb | -0.071 |
| cxcl14 | 0.194 | krt18a.1 | -0.068 |
| LOC110439435 | 0.192 | plecb | -0.068 |
| slc1a2b | 0.191 | actb2 | -0.067 |
| slc25a22b | 0.189 | si:ch211-39i22.1 | -0.067 |
| CABZ01092982.1 | 0.183 | krt18b | -0.067 |
| LO018550.2 | 0.182 | tmsb | -0.065 |
| tfpi2 | 0.180 | capn8 | -0.063 |
| CABZ01043166.1 | 0.178 | tagln2 | -0.062 |
| msx3 | 0.177 | nqo1 | -0.061 |
| si:dkeyp-72a4.1 | 0.176 | krt4 | -0.061 |
| BX547934.2 | 0.176 | wnt2 | -0.060 |
| si:ch211-244b2.4 | 0.176 | krt15 | -0.059 |
| si:ch211-274k16.9 | 0.176 | col17a1b | -0.059 |
| trim101 | 0.174 | negaly6 | -0.058 |
| ahcyl1 | 0.170 | nkx3.2 | -0.057 |
| glula | 0.167 | si:ch211-153b23.5 | -0.055 |
| slc16a1a | 0.166 | txn | -0.055 |
| ncam2 | 0.164 | cyt1 | -0.054 |
| aif1l | 0.163 | cldn8 | -0.054 |
| zwi | 0.162 | cldnh | -0.052 |
| zgc:92275 | 0.162 | hspe1 | -0.051 |
| CU062645.1 | 0.160 | frem3 | -0.051 |
| znf536 | 0.159 | soul2 | -0.051 |
| mb | 0.157 | krt1-c5 | -0.050 |
| kcnq5a | 0.155 | gpc1a | -0.050 |
| nfasca | 0.152 | eif2s2 | -0.050 |
| syngr2b | 0.151 | pvalb8 | -0.050 |
| cahz | 0.151 | wu:fb18f06 | -0.050 |