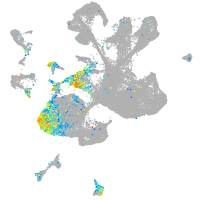
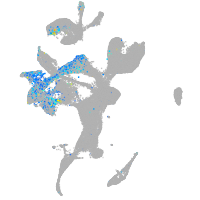
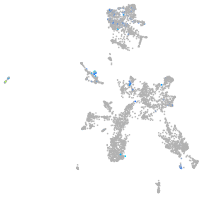
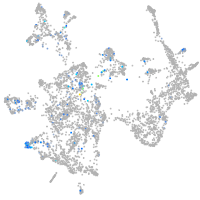
solute carrier family 4 member 4a
ZFIN 
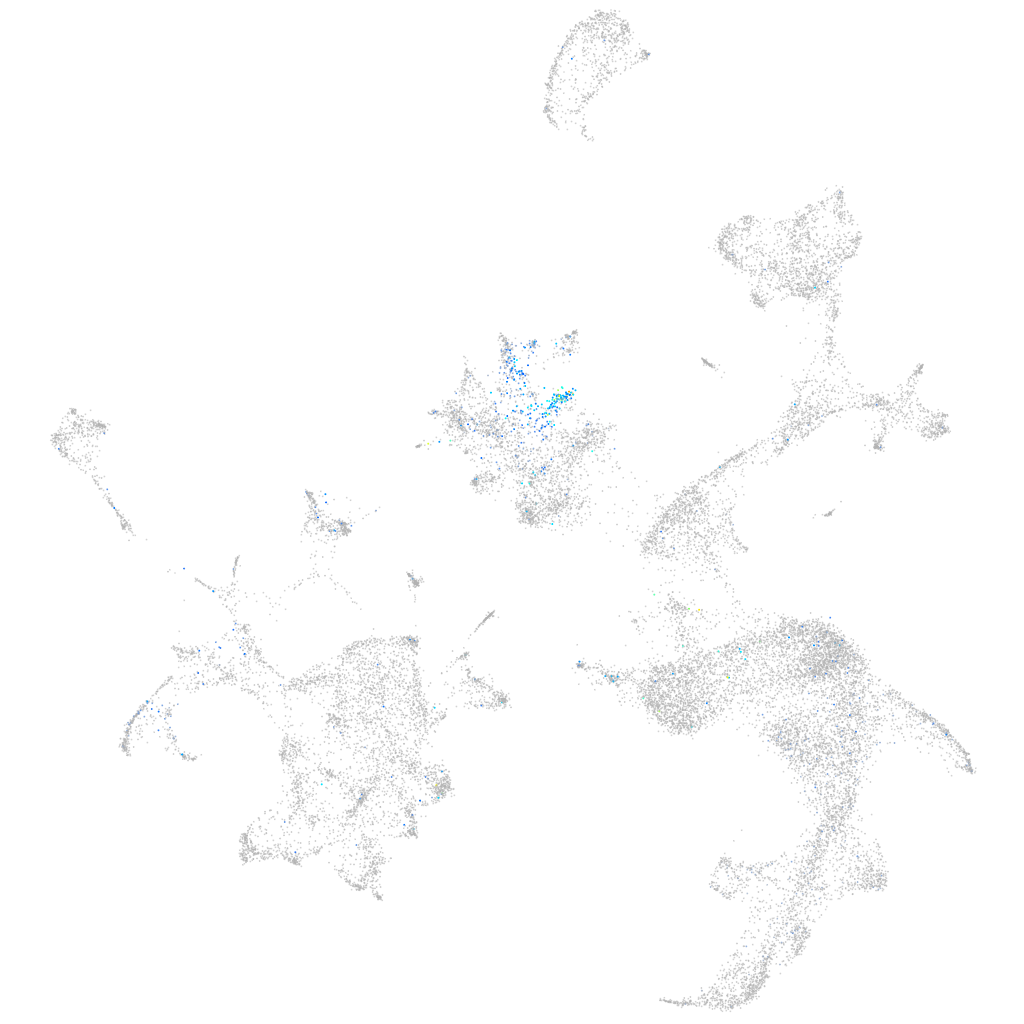
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| slc6a11b | 0.526 | pfn1 | -0.041 |
| hepacama | 0.462 | rac2 | -0.038 |
| atp1a1b | 0.452 | arpc1b | -0.037 |
| slc1a2b | 0.450 | coro1a | -0.036 |
| atp1b4 | 0.440 | pecam1 | -0.035 |
| gpr37l1b | 0.425 | arhgdig | -0.035 |
| si:ch211-66e2.5 | 0.369 | capgb | -0.035 |
| cldn7a | 0.363 | laptm5 | -0.033 |
| FO704813.1 | 0.347 | wasa | -0.033 |
| zgc:165461 | 0.341 | si:dkey-27i16.2 | -0.032 |
| slc6a1b | 0.340 | abcg2a | -0.032 |
| slc1a3b | 0.337 | grap2b | -0.031 |
| sept8b | 0.336 | f13a1b | -0.031 |
| cspg5b | 0.328 | fli1a | -0.031 |
| si:dkey-245n4.2 | 0.326 | si:ch73-234b20.5 | -0.030 |
| fabp7a | 0.319 | FERMT3 (1 of many) | -0.030 |
| slc7a10a | 0.319 | dusp5 | -0.030 |
| gpm6bb | 0.315 | arhgdia | -0.030 |
| hspb15 | 0.312 | sult2st1 | -0.030 |
| ptgdsb.2 | 0.311 | mapre1a | -0.030 |
| eno1b | 0.299 | nrros | -0.029 |
| si:ch73-265d7.2 | 0.291 | fli1b | -0.029 |
| ppap2d | 0.285 | tagln2 | -0.029 |
| mfge8a | 0.283 | wasb | -0.029 |
| efhd1 | 0.281 | dub | -0.029 |
| si:ch211-251b21.1 | 0.275 | inpp5d | -0.029 |
| acbd7 | 0.270 | XLOC-039757 | -0.029 |
| endouc | 0.270 | si:ch211-156j16.1 | -0.028 |
| gfap | 0.264 | hhex | -0.028 |
| wnt7aa | 0.263 | she | -0.028 |
| ptprz1b | 0.258 | dab2 | -0.028 |
| CU467822.1 | 0.255 | cd83 | -0.028 |
| mdkb | 0.253 | si:ch211-102c2.4 | -0.028 |
| mlc1 | 0.251 | etv2 | -0.028 |
| si:ch1073-303k11.2 | 0.246 | abracl | -0.028 |