SIM bHLH transcription factor 2
ZFIN 
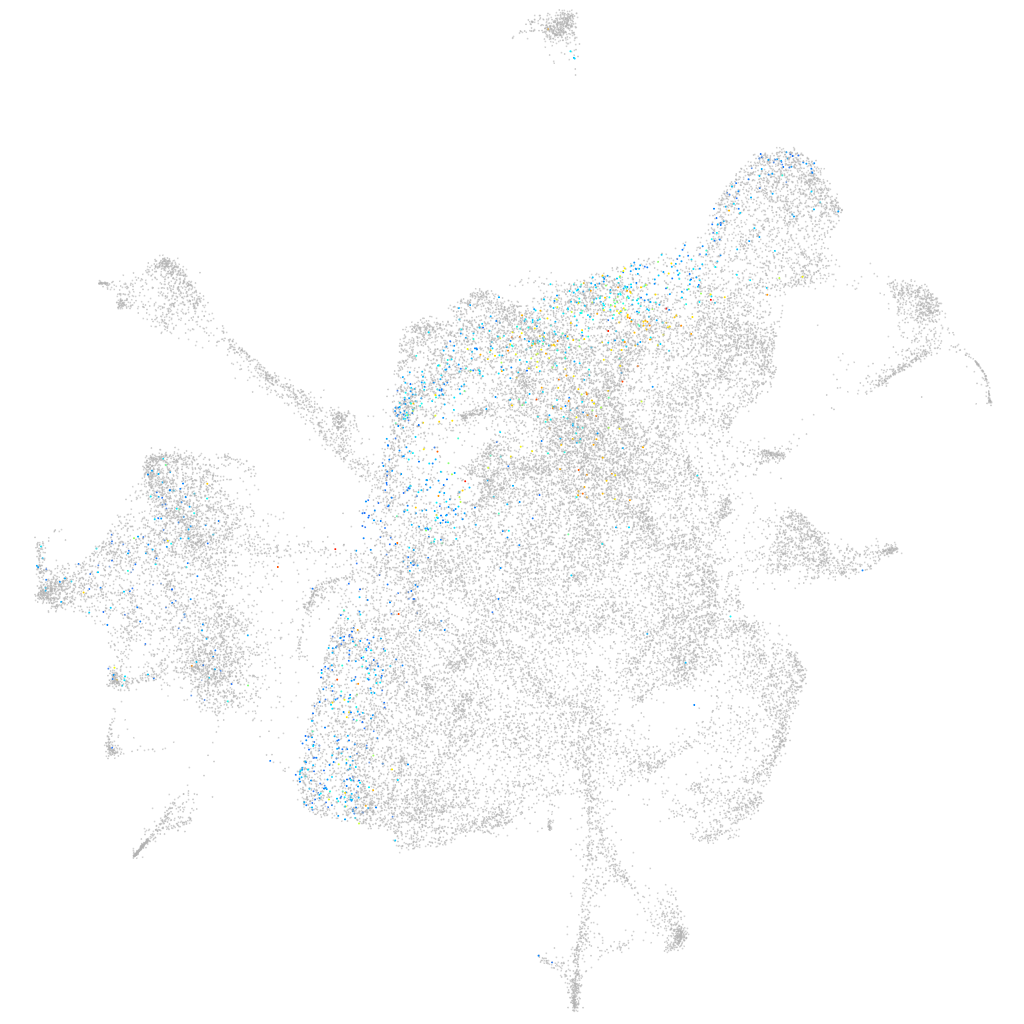
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| sim2.1 | 0.389 | akap12b | -0.055 |
| barx1 | 0.253 | kitlga | -0.052 |
| dlx2a | 0.167 | si:dkey-238c7.16 | -0.049 |
| spon2b | 0.154 | fstl1a | -0.049 |
| dlx1a | 0.152 | ca6 | -0.049 |
| dlx5a | 0.146 | bambia | -0.048 |
| lhx6 | 0.138 | lamc3 | -0.046 |
| cald1b | 0.138 | tmem88b | -0.045 |
| cxcl14 | 0.125 | alx4a | -0.044 |
| foxp2 | 0.123 | tfpia | -0.044 |
| nr2f1b | 0.120 | ucp2 | -0.043 |
| gsc | 0.118 | crabp2a | -0.042 |
| hpgd | 0.113 | tbx15 | -0.042 |
| foxp4 | 0.111 | tuba8l2 | -0.042 |
| epha4b | 0.108 | alcama | -0.041 |
| mab21l2 | 0.105 | msx1b | -0.041 |
| ppp1r3aa | 0.096 | rhag | -0.041 |
| emx2 | 0.094 | pitx2 | -0.040 |
| net1 | 0.090 | tnn | -0.040 |
| ctgfb | 0.089 | zic2a | -0.040 |
| nfic | 0.087 | col4a6 | -0.040 |
| foxl1 | 0.087 | bmp7b | -0.039 |
| sept5a | 0.085 | lfng | -0.039 |
| CU929237.1 | 0.082 | lmx1bb | -0.039 |
| ecrg4a | 0.080 | nid1b | -0.039 |
| sema3ab | 0.080 | hoxb3a | -0.039 |
| tbx22 | 0.079 | gstt1a | -0.039 |
| dlx4b | 0.077 | msx3 | -0.038 |
| sema3c | 0.077 | alx1 | -0.038 |
| vasnb | 0.077 | cxadr | -0.038 |
| hyal4 | 0.076 | XLOC-042222 | -0.037 |
| fgfrl1a | 0.076 | col4a5 | -0.037 |
| foxc1b | 0.075 | hoxd9a | -0.036 |
| nfixb | 0.073 | cdon | -0.036 |
| dlx4a | 0.073 | cavin2b | -0.036 |