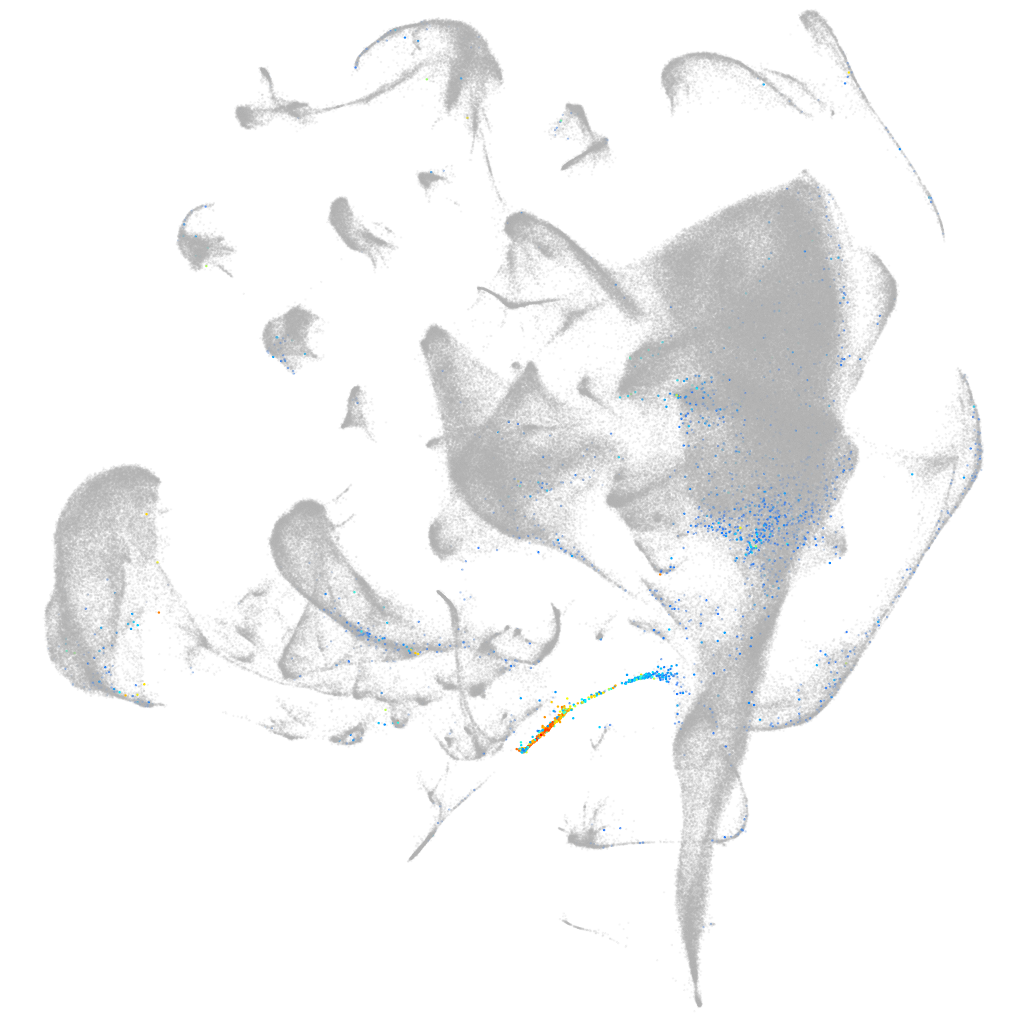
si:dkey-239j18.3
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature 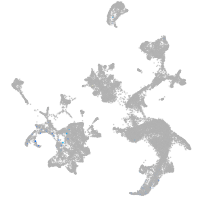 |
pronephros 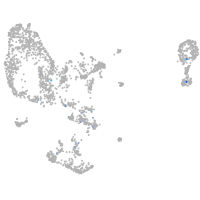 |
neural |
spinal cord / glia |
eye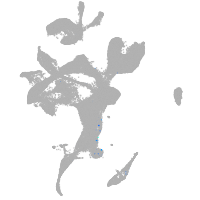 |
taste / olfactory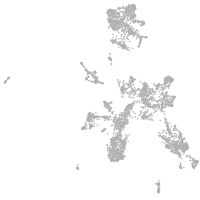 |
otic / lateral line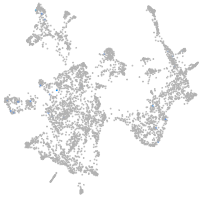 |
epidermis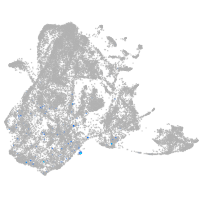 |
periderm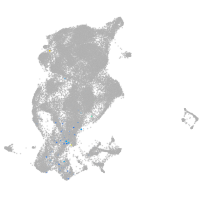 |
fin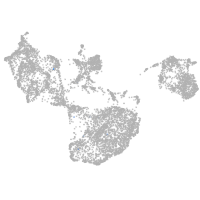 |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| LOC108180223 | 0.497 | pvalb1 | -0.031 |
| he1.2 | 0.467 | sod1 | -0.031 |
| ctslb | 0.435 | pvalb2 | -0.029 |
| si:dkey-269i1.4 | 0.429 | prdx2 | -0.027 |
| zgc:174153 | 0.421 | gapdhs | -0.026 |
| si:dkey-26g8.4 | 0.420 | adh5 | -0.025 |
| si:dkey-239j18.2 | 0.410 | ldhba | -0.025 |
| si:dkey-26g8.5 | 0.357 | stmn1b | -0.025 |
| he1.1 | 0.355 | gpm6aa | -0.024 |
| wu:fa26c03 | 0.355 | krtt1c19e | -0.024 |
| sb:cb470 | 0.336 | ndrg2 | -0.024 |
| si:dkey-269i1.3 | 0.336 | fxyd6l | -0.023 |
| tmprss9 | 0.304 | gpm6ab | -0.023 |
| bhlha15 | 0.254 | rbp4 | -0.023 |
| vtcn1 | 0.247 | ahcy | -0.022 |
| timp4.1 | 0.201 | ccni | -0.021 |
| foxe3 | 0.190 | celf2 | -0.021 |
| mgp | 0.190 | si:dkey-33c14.3 | -0.021 |
| scin | 0.165 | ckbb | -0.020 |
| xkr9 | 0.157 | gng3 | -0.020 |
| slc30a8 | 0.153 | ip6k2a | -0.020 |
| klf17 | 0.144 | kdm6bb | -0.020 |
| b3gnt5b | 0.143 | prdx6 | -0.020 |
| si:ch211-163l21.8 | 0.142 | rtn1a | -0.020 |
| svopl | 0.129 | si:dkey-16p21.8 | -0.020 |
| fmn1 | 0.127 | tcnbb | -0.020 |
| lad1 | 0.125 | tuba1c | -0.020 |
| mctp2b | 0.123 | wu:fa03e10 | -0.020 |
| gpr160 | 0.118 | zgc:162944 | -0.020 |
| sytl4 | 0.118 | atp6v0cb | -0.019 |
| pitx2 | 0.115 | cap1 | -0.019 |
| zgc:158403 | 0.111 | cbx1b | -0.019 |
| tram1 | 0.109 | cotl1 | -0.019 |
| rgs2 | 0.106 | tnrc6c1 | -0.019 |
| fkbp11 | 0.105 | vdac3 | -0.019 |