si:dkey-239j18.3
ZFIN 
Other cell groups


all cells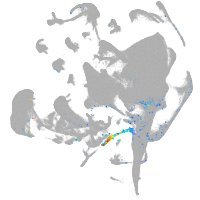 |
blastomeres |
PGCs |
endoderm |
axial mesoderm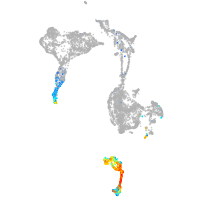 |
muscle 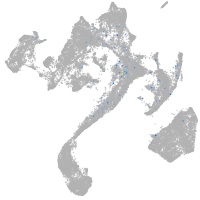 |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature 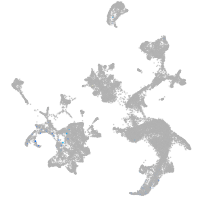 |
pronephros 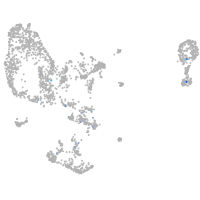 |
neural |
spinal cord / glia |
eye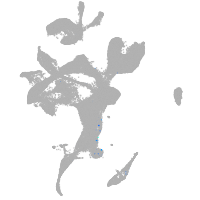 |
taste / olfactory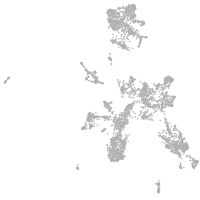 |
otic / lateral line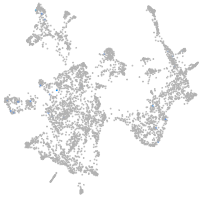 |
epidermis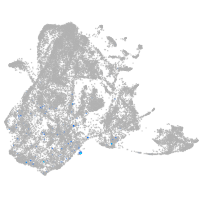 |
periderm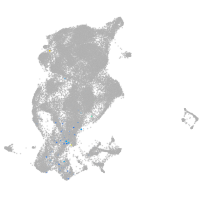 |
fin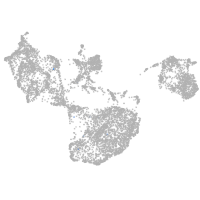 |
ionocytes / mucous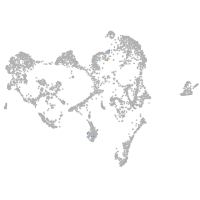 |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| zgc:174153 | 0.240 | ckbb | -0.033 |
| si:dkey-239j18.2 | 0.236 | COX3 | -0.032 |
| ctslb | 0.236 | mt-co2 | -0.031 |
| he1.2 | 0.232 | gapdhs | -0.030 |
| si:dkey-26g8.5 | 0.216 | atp6v0cb | -0.029 |
| wu:fa26c03 | 0.216 | snap25b | -0.028 |
| si:busm1-160c18.6 | 0.211 | rnasekb | -0.028 |
| si:dkey-269i1.3 | 0.190 | mt-atp6 | -0.027 |
| si:dkey-269i1.4 | 0.187 | pvalb2 | -0.024 |
| he1.1 | 0.173 | syt5b | -0.024 |
| scarf1 | 0.151 | sypb | -0.023 |
| si:dkey-26g8.4 | 0.142 | pvalb1 | -0.022 |
| gypc | 0.101 | atpv0e2 | -0.021 |
| arhgef40 | 0.099 | gpm6aa | -0.021 |
| slc29a1b | 0.088 | ndrg4 | -0.021 |
| si:dkey-24c2.9 | 0.082 | rs1a | -0.021 |
| ecscr | 0.077 | mdkb | -0.021 |
| cxcr3.3 | 0.076 | tpi1b | -0.020 |
| map1lc3cl | 0.072 | atp6v1e1b | -0.020 |
| si:dkey-32n7.4 | 0.068 | atp1a3a | -0.020 |
| si:dkey-238i5.3 | 0.062 | gpm6ab | -0.020 |
| CR769772.4 | 0.060 | calb2b | -0.020 |
| si:ch211-248e11.2 | 0.059 | otx5 | -0.020 |
| fgd5a | 0.058 | mpp6b | -0.020 |
| trpm4a | 0.057 | anp32e | -0.019 |
| acvrl1 | 0.055 | crx | -0.019 |
| atr | 0.054 | calm1a | -0.019 |
| col8a1a | 0.054 | vamp2 | -0.019 |
| sox21b | 0.050 | atp1b2a | -0.018 |
| si:ch73-389b16.1 | 0.050 | rtn1a | -0.018 |
| or128-1 | 0.047 | hmgn6 | -0.018 |
| myzap | 0.046 | gng13b | -0.018 |
| paics | 0.045 | stmn1b | -0.018 |
| hoxb6b | 0.045 | pcbp3 | -0.017 |
| XLOC-031367 | 0.044 | sh3gl2a | -0.017 |




