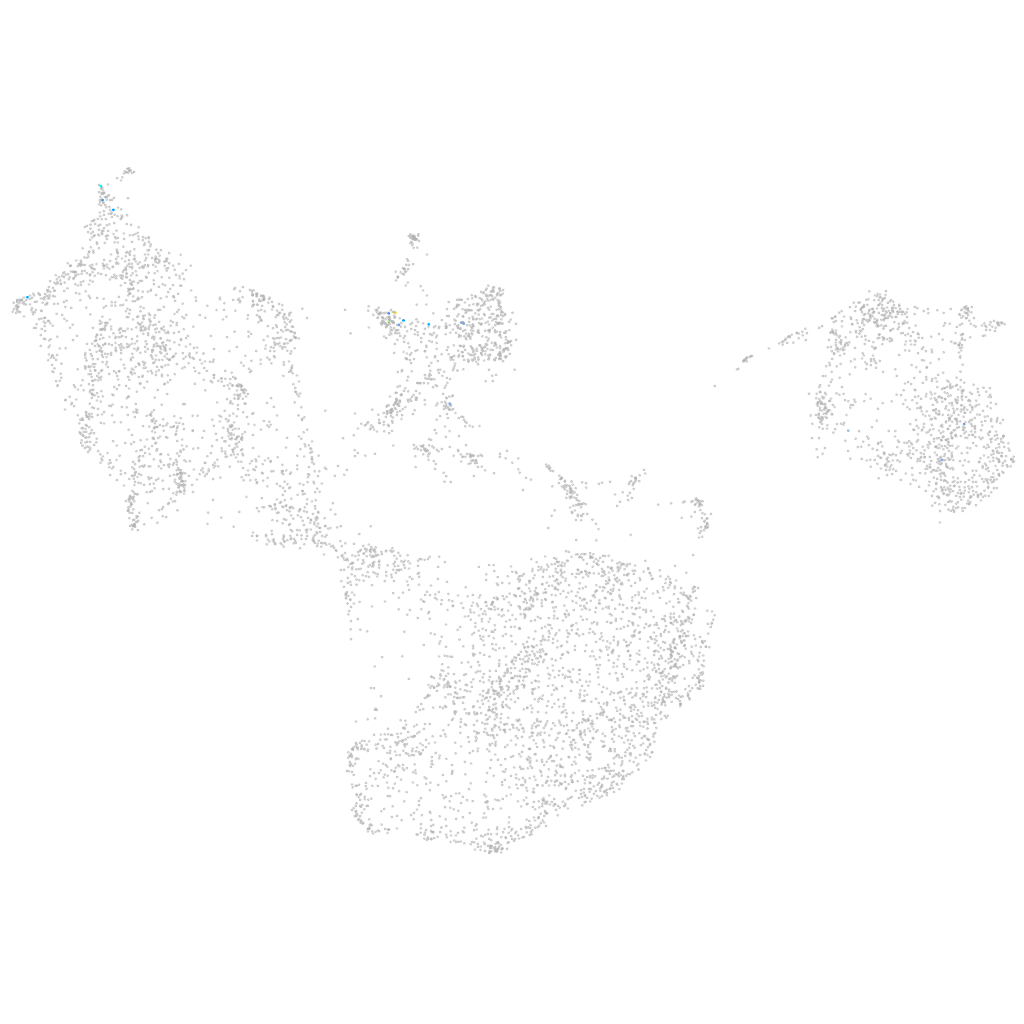
si:dkey-165a24.9
ZFIN 
Other cell groups


all cells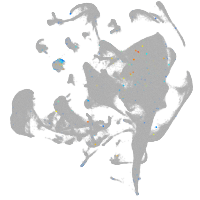 |
blastomeres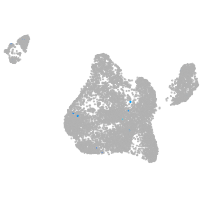 |
PGCs |
endoderm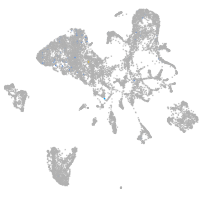 |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros 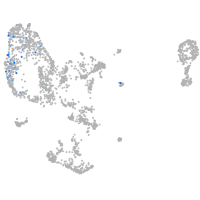 |
neural |
spinal cord / glia |
eye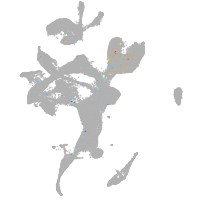 |
taste / olfactory |
otic / lateral line |
epidermis |
periderm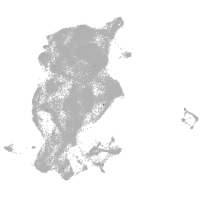 |
fin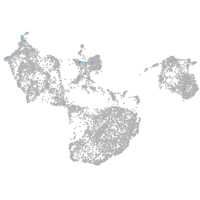 |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| si:ch211-57i17.2 | 0.417 | krt4 | -0.044 |
| si:dkey-69c1.1 | 0.370 | cfl1l | -0.044 |
| abi3a | 0.354 | s100a10b | -0.042 |
| si:ch73-22o18.1 | 0.331 | cldni | -0.041 |
| si:ch211-276c2.2 | 0.324 | tmsb1 | -0.041 |
| si:ch211-117n7.7 | 0.315 | cldn1 | -0.041 |
| galnt16 | 0.286 | epcam | -0.040 |
| tgfa | 0.283 | pfn1 | -0.039 |
| LAMP5 | 0.263 | ier2b | -0.039 |
| gabrb2l | 0.258 | cyt1 | -0.038 |
| LOC110438259 | 0.258 | anxa1a | -0.037 |
| baiap2b | 0.258 | ahnak | -0.036 |
| ahr1b | 0.256 | thy1 | -0.036 |
| ephb6 | 0.256 | col18a1a | -0.036 |
| RIC3 | 0.250 | spaca4l | -0.036 |
| FITM1 (1 of many) | 0.248 | apoeb | -0.035 |
| pimr80 | 0.247 | tln1 | -0.035 |
| ccdc88aa | 0.246 | pak2a | -0.035 |
| pimr83 | 0.243 | rac2 | -0.034 |
| opn1mw3 | 0.241 | myh9a | -0.034 |
| ush1ga | 0.239 | tmsb4x | -0.034 |
| zgc:162324 | 0.235 | eno3 | -0.034 |
| LOC792447 | 0.235 | bcam | -0.034 |
| fam43b | 0.233 | ak1 | -0.033 |
| zgc:56622 | 0.231 | krt8 | -0.033 |
| lactbl1a | 0.230 | si:dkey-102c8.3 | -0.033 |
| hmx1 | 0.225 | ecrg4b | -0.033 |
| si:ch211-232m10.6 | 0.224 | pycard | -0.033 |
| BX927308.2 | 0.223 | wu:fb18f06 | -0.033 |
| CABZ01087025.3 | 0.222 | dnase1l4.1 | -0.033 |
| apof | 0.219 | epgn | -0.032 |
| cntn4 | 0.214 | cap1 | -0.032 |
| CR847537.1 | 0.210 | zgc:92380 | -0.032 |
| si:dkey-9i23.8 | 0.210 | mcl1b | -0.032 |
| zgc:103625 | 0.209 | zgc:158343 | -0.032 |