si:dkey-165a24.9
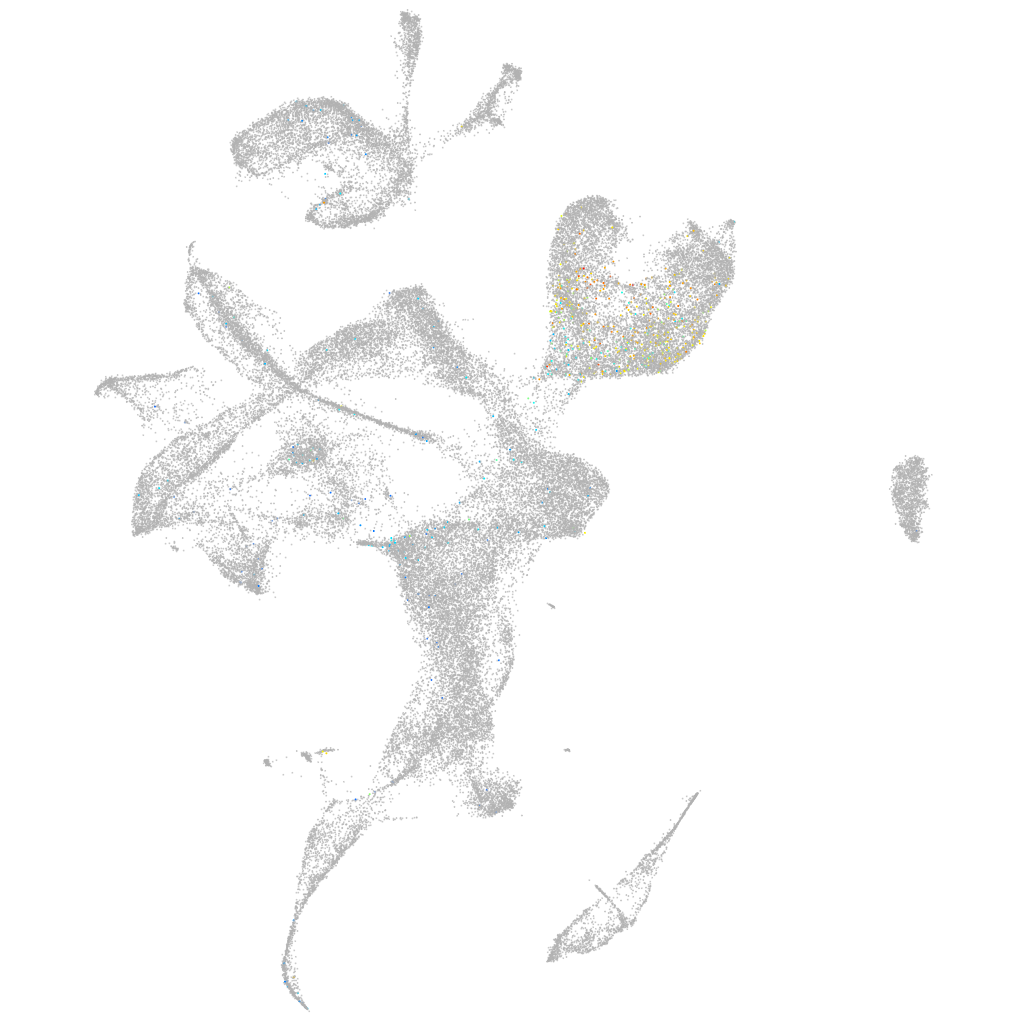
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros 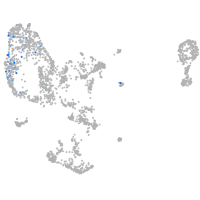 |
neural |
spinal cord / glia |
eye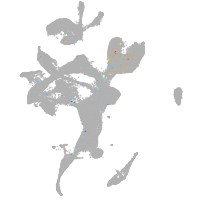 |
taste / olfactory |
otic / lateral line |
epidermis |
periderm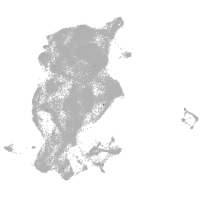 |
fin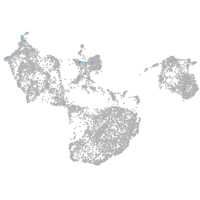 |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| gnb3a | 0.128 | tma7 | -0.061 |
| gng13b | 0.115 | stmn1a | -0.060 |
| nsg2 | 0.114 | anp32b | -0.059 |
| hunk | 0.109 | pcna | -0.057 |
| dscaml1 | 0.108 | zgc:56493 | -0.056 |
| samsn1a | 0.106 | fabp7a | -0.056 |
| pcp4l1 | 0.106 | msi1 | -0.054 |
| bcl2l10 | 0.103 | tuba8l4 | -0.054 |
| mpp6b | 0.102 | rpl5a | -0.053 |
| vsx1 | 0.101 | banf1 | -0.053 |
| tp53inp1 | 0.100 | dynll1 | -0.053 |
| chst1 | 0.098 | selenoh | -0.053 |
| scrt2 | 0.096 | apex1 | -0.053 |
| ddah2 | 0.091 | si:ch73-281n10.2 | -0.053 |
| mdkb | 0.090 | COX5B | -0.052 |
| ndrg4 | 0.090 | dut | -0.052 |
| btg1 | 0.089 | rpl4 | -0.051 |
| neurod4 | 0.089 | hspe1 | -0.051 |
| gpm6ab | 0.088 | snrpd1 | -0.050 |
| nova2 | 0.087 | eif2s2 | -0.050 |
| crx | 0.087 | snrpb | -0.050 |
| prox1a | 0.086 | nasp | -0.050 |
| rorab | 0.086 | atp5if1a | -0.050 |
| marcksl1a | 0.085 | ahcy | -0.050 |
| ppp1r1b | 0.085 | snu13b | -0.049 |
| otx5 | 0.085 | nop10 | -0.049 |
| syt5b | 0.085 | snrpe | -0.049 |
| SEMA4F | 0.084 | dek | -0.049 |
| bhlhe23 | 0.082 | rrm1 | -0.049 |
| epb41a | 0.080 | ccnd1 | -0.049 |
| daam1a | 0.079 | cbx3a | -0.048 |
| zgc:100920 | 0.079 | nutf2l | -0.048 |
| eml1 | 0.078 | ranbp1 | -0.048 |
| ddit3 | 0.078 | nop58 | -0.048 |
| slitrk6 | 0.077 | fen1 | -0.048 |