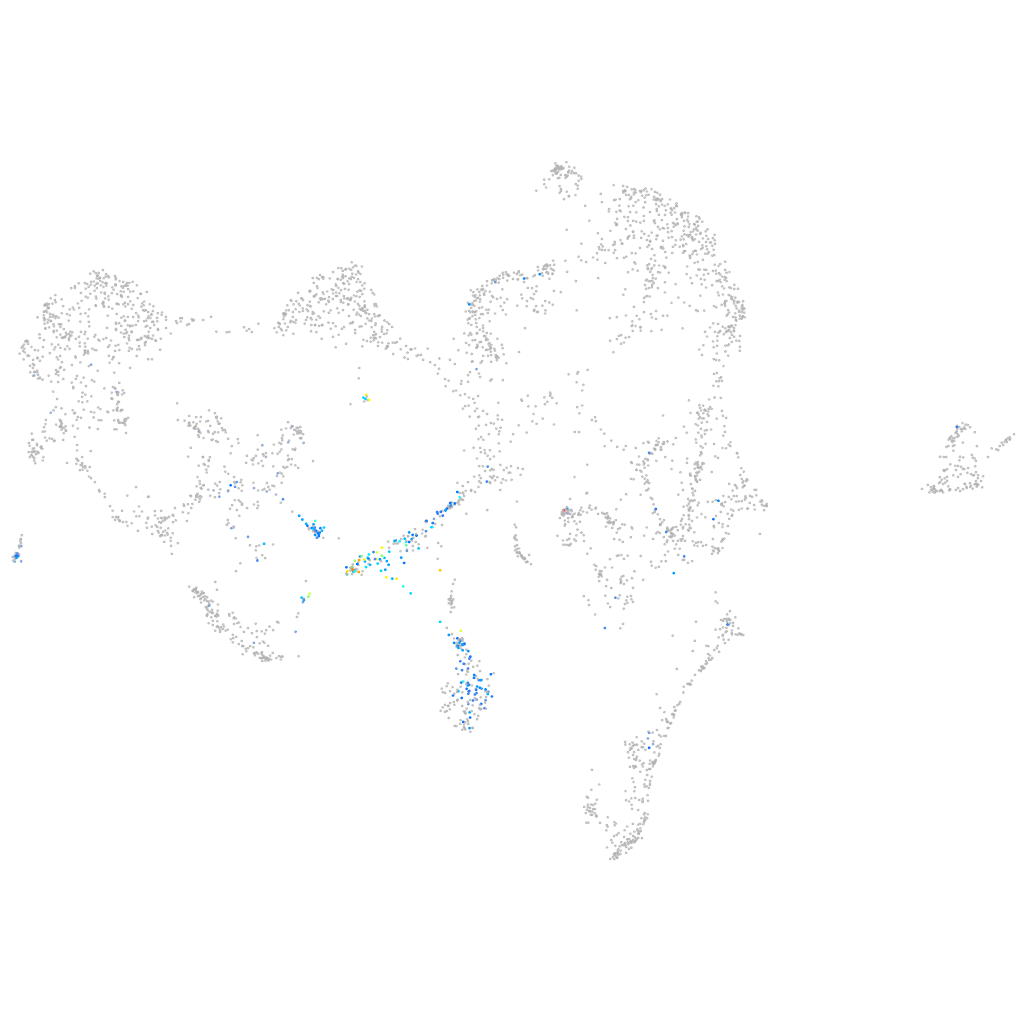
si:dkey-14d8.20
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| ceacam1 | 0.578 | krt91 | -0.182 |
| si:dkey-192d15.2 | 0.572 | BX000438.2 | -0.168 |
| zgc:193726 | 0.548 | rpl37 | -0.160 |
| ly6m2 | 0.528 | si:dkey-151g10.6 | -0.154 |
| slc6a6b | 0.527 | junba | -0.153 |
| eef2l2 | 0.525 | eef1a1l1 | -0.138 |
| rhcgb | 0.519 | spaca4l | -0.137 |
| cox5b2 | 0.517 | actb1 | -0.137 |
| ca15a | 0.514 | hmgn2 | -0.136 |
| slc4a1b | 0.508 | lgals3b | -0.136 |
| atp6v0a1a | 0.508 | rplp1 | -0.134 |
| atp6v1aa | 0.493 | aldob | -0.131 |
| clcn2b | 0.489 | agr2 | -0.130 |
| serpinc1 | 0.477 | hmgn3 | -0.128 |
| atp6ap2 | 0.454 | krt5 | -0.128 |
| atp6v0ca | 0.442 | her6 | -0.126 |
| atp6v1e1b | 0.441 | malb | -0.125 |
| atp6ap1b | 0.441 | dap1b | -0.124 |
| atp6v1g1 | 0.436 | fa2h | -0.123 |
| atp6v1ba | 0.434 | rps21 | -0.120 |
| rnaseka | 0.430 | slc38a5b | -0.118 |
| slc31a2 | 0.429 | eif4ebp3l | -0.118 |
| atp6v1c1b | 0.422 | s100v1 | -0.115 |
| atp6v1d | 0.417 | si:dkey-65b12.6 | -0.113 |
| foxi3a | 0.414 | muc5.1 | -0.112 |
| ca2 | 0.412 | rpl36 | -0.111 |
| CDK18 | 0.410 | rpl21 | -0.111 |
| atp6v0d1 | 0.410 | fxyd6l | -0.110 |
| etf1a | 0.397 | rps20 | -0.110 |
| chp1 | 0.393 | nupr1b | -0.109 |
| chst2a | 0.389 | b3gnt7 | -0.109 |
| atp6v0b | 0.388 | eef1db | -0.108 |
| atp6v1h | 0.388 | her9 | -0.108 |
| gb:bc139872 | 0.375 | sdc4 | -0.108 |
| si:dkeyp-72e1.7 | 0.375 | rps24 | -0.108 |