si:ch211-197h24.8
ZFIN 
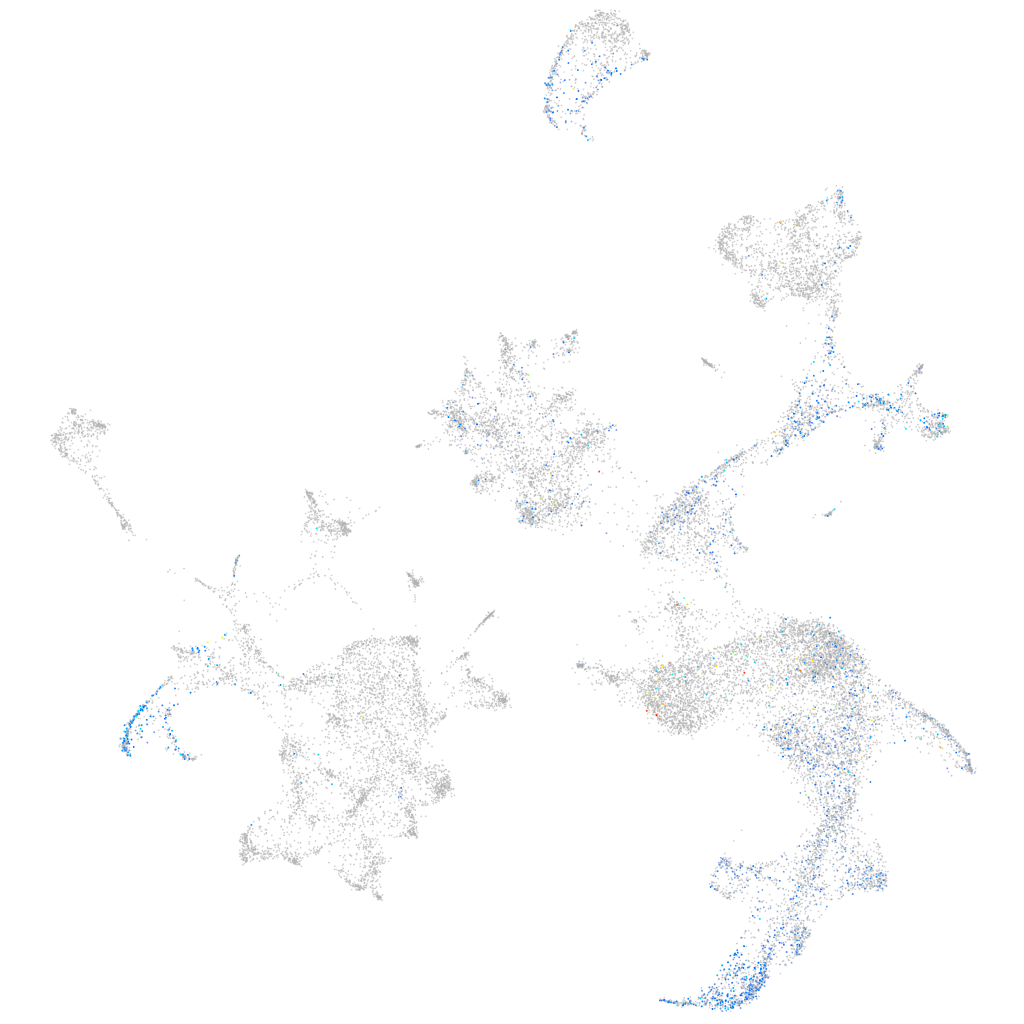
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| cpox | 0.224 | marcksl1b | -0.122 |
| urod | 0.207 | krt8 | -0.109 |
| si:dkey-261j4.4 | 0.207 | ctsla | -0.106 |
| hmbsa | 0.207 | marcksl1a | -0.105 |
| cldng | 0.202 | cd81a | -0.103 |
| wu:fb97g03 | 0.201 | krt18a.1 | -0.102 |
| znfl2a | 0.196 | fxyd6l | -0.101 |
| blf | 0.191 | cpn1 | -0.100 |
| klf17 | 0.189 | cdh5 | -0.097 |
| alad | 0.180 | akap12b | -0.093 |
| hbbe3 | 0.175 | tmsb4x | -0.092 |
| hmbsb | 0.170 | cldn5b | -0.091 |
| drl | 0.169 | vdac3 | -0.090 |
| glrx5 | 0.168 | si:ch211-156j16.1 | -0.089 |
| abcb10 | 0.166 | dap1b | -0.088 |
| si:dkey-261j4.3 | 0.164 | vat1 | -0.087 |
| gata1a | 0.163 | nova2 | -0.087 |
| hdr | 0.160 | tie1 | -0.087 |
| abce1 | 0.151 | plvapb | -0.087 |
| txn | 0.150 | clec14a | -0.086 |
| csrnp1a | 0.147 | fstl1b | -0.086 |
| bcas2 | 0.143 | jpt1b | -0.086 |
| si:ch73-299h12.2 | 0.143 | fabp11a | -0.086 |
| tfr1a | 0.141 | igfbp7 | -0.086 |
| slc38a5a | 0.140 | si:ch211-145b13.6 | -0.085 |
| mllt3 | 0.140 | egfl7 | -0.084 |
| tmem14ca | 0.138 | myct1a | -0.084 |
| myb | 0.136 | col4a1 | -0.084 |
| XLOC-001964 | 0.134 | ramp2 | -0.083 |
| anp32b | 0.134 | she | -0.083 |
| susd1 | 0.133 | id1 | -0.083 |
| suv39h1b | 0.132 | si:dkey-7j14.6 | -0.083 |
| phlda2 | 0.131 | cnn3a | -0.082 |
| blvra | 0.130 | ecscr | -0.082 |
| slc11a2 | 0.129 | pecam1 | -0.081 |