si:ch211-147g22.4
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis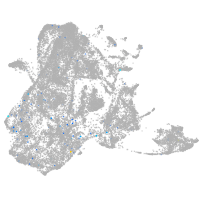 |
periderm |
fin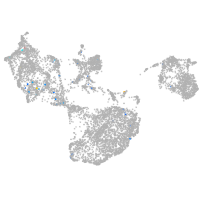 |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| egr3 | 1.000 | si:ch211-288g17.3 | -0.279 |
| LO017816.1 | 1.000 | rps18 | -0.202 |
| pstpip1a | 1.000 | rhoaa | -0.186 |
| tpra1 | 1.000 | pno1 | -0.179 |
| slc6a6a | 0.780 | nucks1a | -0.172 |
| dub | 0.747 | COX3 | -0.169 |
| aass | 0.724 | srrm1 | -0.168 |
| adcy8 | 0.724 | hnrnpaba | -0.168 |
| antxr1d | 0.724 | hnrnpa0a | -0.168 |
| BX284635.1 | 0.724 | fbl | -0.167 |
| BX510649.1 | 0.724 | mt-co2 | -0.166 |
| BX936461.1 | 0.724 | gar1 | -0.163 |
| caln1 | 0.724 | zgc:112356 | -0.160 |
| CR932077.1 | 0.724 | hp1bp3 | -0.157 |
| CR933528.1 | 0.724 | pcna | -0.156 |
| CU302321.1 | 0.724 | syncrip | -0.152 |
| fabp10b | 0.724 | nasp | -0.152 |
| FP103004.1 | 0.724 | pole4 | -0.152 |
| gcgra | 0.724 | srrm2 | -0.151 |
| gpr155b | 0.724 | kri1 | -0.150 |
| grin3ba | 0.724 | htatsf1 | -0.150 |
| hoxc6a | 0.724 | rnf2 | -0.148 |
| ksr2 | 0.724 | zc3h18 | -0.147 |
| LOC100538221 | 0.724 | dynll2a | -0.147 |
| LOC110439953 | 0.724 | top1l | -0.147 |
| lrfn2b | 0.724 | nap1l1 | -0.146 |
| pnp4a | 0.724 | srsf3a | -0.145 |
| prdm1c | 0.724 | llph | -0.145 |
| si:ch211-197g15.8 | 0.724 | ncl | -0.145 |
| si:dkey-18a10.3 | 0.724 | anp32e | -0.144 |
| slc17a7a | 0.724 | dek | -0.144 |
| tmem176l.2 | 0.724 | mki67 | -0.143 |
| XLOC-008494 | 0.724 | cdv3 | -0.143 |
| XLOC-009629 | 0.724 | cdca7b | -0.142 |
| XLOC-014155 | 0.724 | adssl | -0.141 |