Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia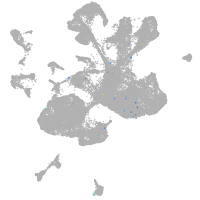 |
eye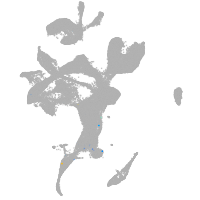 |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| tgm2a | 0.085 | ckbb | -0.015 |
| rtn2b | 0.078 | epcam | -0.015 |
| mybpc2b | 0.076 | icn | -0.015 |
| cavin4a | 0.073 | krt8 | -0.015 |
| myoz1b | 0.073 | selenow1 | -0.015 |
| hhatla | 0.072 | tmsb1 | -0.015 |
| cavin4b | 0.071 | cfl1l | -0.014 |
| tnnt3a | 0.071 | cyt1 | -0.014 |
| casq1a | 0.070 | gapdhs | -0.014 |
| limd1b | 0.070 | krt4 | -0.014 |
| zgc:158296 | 0.070 | s100a10b | -0.014 |
| cdnf | 0.069 | ywhaz | -0.014 |
| prr33 | 0.069 | zgc:162730 | -0.014 |
| myhc4 | 0.068 | anxa1a | -0.013 |
| CABZ01072309.1 | 0.067 | anxa4 | -0.013 |
| CABZ01078594.1 | 0.067 | atp6v0cb | -0.013 |
| cacna1sb | 0.067 | cap1 | -0.013 |
| cx39.9 | 0.066 | cd9b | -0.013 |
| myom2a | 0.066 | cebpb | -0.013 |
| xirp2b | 0.066 | cotl1 | -0.013 |
| znf648 | 0.066 | gpm6ab | -0.013 |
| pgam2 | 0.066 | krtt1c19e | -0.013 |
| trdn | 0.066 | pfn1 | -0.013 |
| casq2 | 0.065 | s100v2 | -0.013 |
| dhrs7cb | 0.065 | stmn1b | -0.013 |
| ldb3b | 0.065 | sult6b1 | -0.013 |
| myhz1.3 | 0.065 | calm1a | -0.013 |
| myom1a | 0.065 | anxa2a | -0.012 |
| si:ch1073-100f3.2 | 0.065 | arhgdig | -0.012 |
| casq1b | 0.064 | arhgef5 | -0.012 |
| CR376766.2 | 0.064 | atp6v1e1b | -0.012 |
| myoz2a | 0.064 | cebpd | -0.012 |
| ryr1b | 0.064 | dnase1l4.1 | -0.012 |
| srl | 0.064 | fbp1a | -0.012 |
| tmem182a | 0.064 | gng3 | -0.012 |




