short stature homeobox
ZFIN 
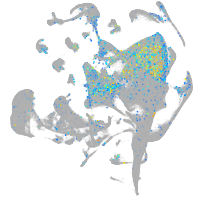
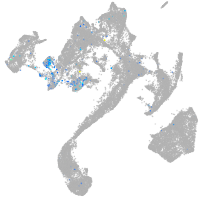
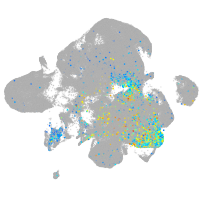
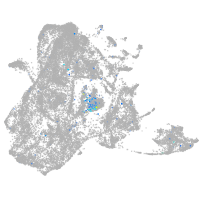
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| shox2 | 0.265 | tspan36 | -0.089 |
| adcyap1b | 0.257 | smim29 | -0.079 |
| tfap2d | 0.241 | pttg1ipb | -0.077 |
| elavl3 | 0.235 | syngr1a | -0.077 |
| barhl1a | 0.227 | gpr143 | -0.076 |
| stmn1b | 0.223 | cst14a.2 | -0.074 |
| nrn1a | 0.217 | paics | -0.072 |
| ebf3a | 0.217 | rnaseka | -0.071 |
| lmo3 | 0.213 | trpm1b | -0.069 |
| tmeff1b | 0.210 | rabl6b | -0.067 |
| gpm6aa | 0.210 | slc45a2 | -0.066 |
| tmsb | 0.209 | rab32a | -0.066 |
| fez1 | 0.205 | bace2 | -0.065 |
| gng2 | 0.204 | agtrap | -0.065 |
| pou4f2 | 0.203 | pcbd1 | -0.065 |
| nova2 | 0.200 | rab38 | -0.064 |
| runx1t1 | 0.197 | gstp1 | -0.063 |
| tubb5 | 0.194 | atox1 | -0.063 |
| lhx9 | 0.194 | gmps | -0.063 |
| tuba1c | 0.193 | impdh1b | -0.060 |
| ptmaa | 0.192 | glulb | -0.060 |
| rtn1a | 0.187 | zgc:165573 | -0.060 |
| gng3 | 0.186 | sdc4 | -0.060 |
| hmgb3a | 0.185 | eef1a1l1 | -0.058 |
| chd4a | 0.185 | slc38a5b | -0.058 |
| cdk5r1b | 0.183 | zgc:110239 | -0.057 |
| BX323080.1 | 0.183 | eno3 | -0.057 |
| dync1i1 | 0.182 | ahnak | -0.057 |
| barhl1b | 0.180 | sypl2b | -0.057 |
| fyna | 0.180 | ctsd | -0.056 |
| LOC100000890 | 0.180 | si:dkey-204f11.64 | -0.056 |
| bcl11ba | 0.180 | tspan10 | -0.056 |
| mab21l1 | 0.180 | rpl5a | -0.055 |
| tox | 0.179 | mitfa | -0.054 |
| ebf1a | 0.179 | qdpra | -0.054 |