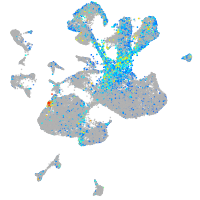
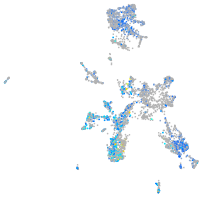
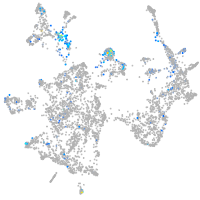
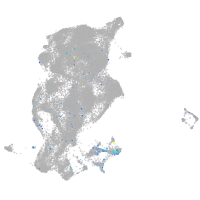
septin 5a
ZFIN 
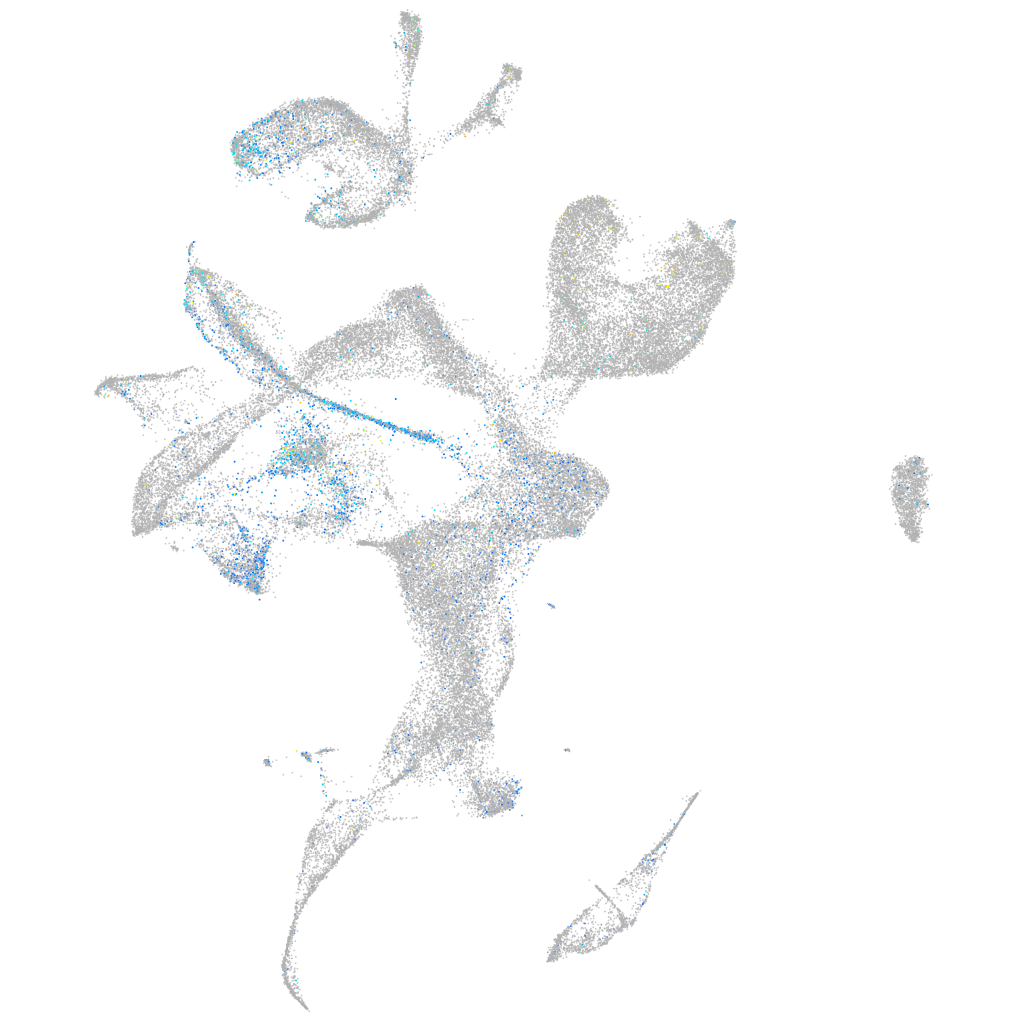
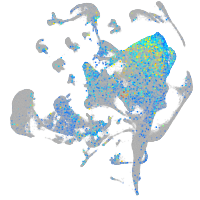
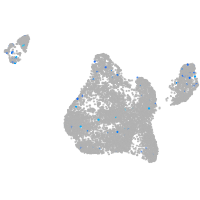
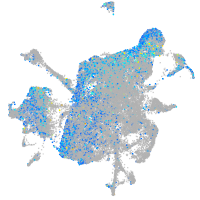
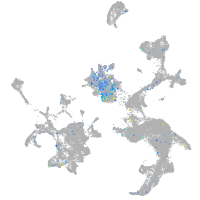
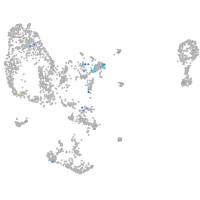
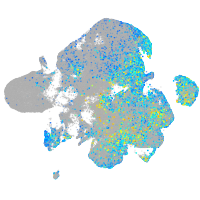
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| elavl3 | 0.188 | hmgb2a | -0.082 |
| mllt11 | 0.174 | anp32e | -0.055 |
| cxcr4b | 0.172 | crx | -0.054 |
| zgc:65894 | 0.151 | otx5 | -0.052 |
| tubb5 | 0.150 | syt5b | -0.047 |
| stmn2a | 0.147 | nop58 | -0.045 |
| klf7b | 0.143 | fbl | -0.045 |
| tmsb | 0.141 | vsx2 | -0.045 |
| stmn1b | 0.139 | dkc1 | -0.043 |
| snap25a | 0.139 | ndrg1b | -0.043 |
| rtn1a | 0.139 | chaf1a | -0.042 |
| ebf2 | 0.138 | ccnd1 | -0.041 |
| ywhag2 | 0.136 | rs1a | -0.040 |
| gap43 | 0.135 | mki67 | -0.040 |
| elavl4 | 0.133 | vsx1 | -0.040 |
| rtn1b | 0.131 | rsl1d1 | -0.039 |
| alcama | 0.130 | nop2 | -0.039 |
| nrn1a | 0.124 | lig1 | -0.039 |
| tuba1c | 0.124 | nop56 | -0.039 |
| stxbp1a | 0.123 | ddah2 | -0.039 |
| irx1a | 0.120 | pcna | -0.038 |
| eno2 | 0.120 | mcm6 | -0.038 |
| gng2 | 0.119 | npm1a | -0.037 |
| si:dkeyp-75h12.5 | 0.119 | atic | -0.037 |
| rbpms2b | 0.119 | lyar | -0.036 |
| syn2a | 0.118 | mcm5 | -0.036 |
| gng3 | 0.118 | mcm3 | -0.036 |
| zgc:158291 | 0.116 | dek | -0.036 |
| tmsb4x | 0.116 | snu13b | -0.036 |
| dpysl3 | 0.115 | CU929070.1 | -0.035 |
| sncb | 0.114 | ddx18 | -0.035 |
| adcyap1b | 0.112 | dnmbp | -0.035 |
| rbfox2 | 0.112 | cdca7a | -0.035 |
| nhlh2 | 0.112 | gnl3 | -0.035 |
| gng7 | 0.112 | gnb3a | -0.034 |