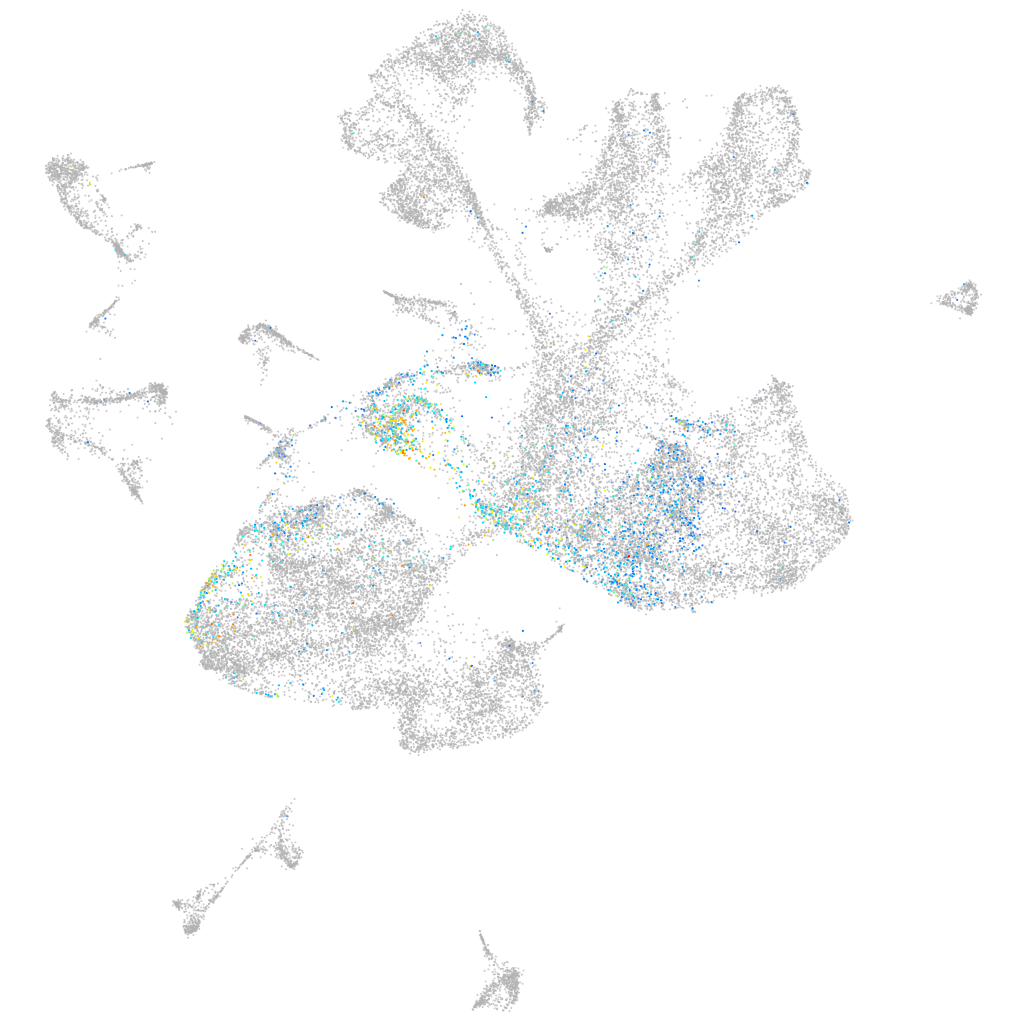
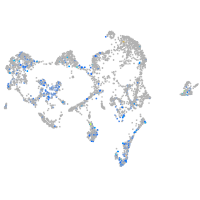
semaphorin 4e
ZFIN 
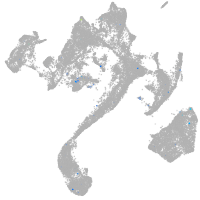
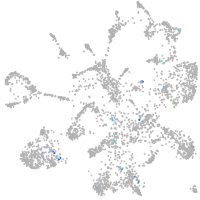
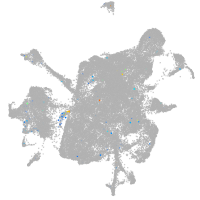
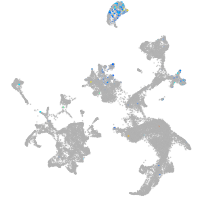
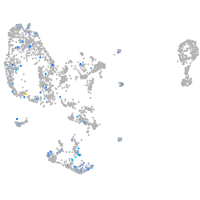
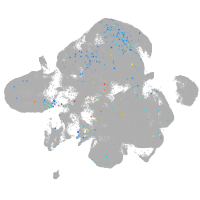
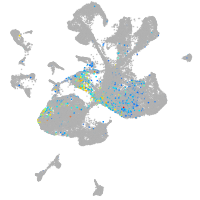
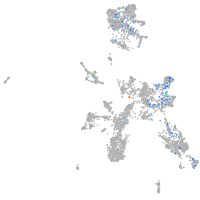
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| si:ch211-251b21.1 | 0.277 | elavl3 | -0.111 |
| endouc | 0.237 | si:ch211-222l21.1 | -0.107 |
| plecb | 0.235 | ptmaa | -0.102 |
| col4a5 | 0.232 | ptmab | -0.098 |
| gfap | 0.232 | h3f3d | -0.096 |
| slc6a9 | 0.221 | stmn1b | -0.090 |
| atp1a1b | 0.219 | myt1b | -0.087 |
| cav1 | 0.217 | gng3 | -0.084 |
| cldn7a | 0.216 | rtn1a | -0.084 |
| si:dkey-245n4.2 | 0.216 | tubb5 | -0.082 |
| wu:fi04e12 | 0.213 | stxbp1a | -0.081 |
| prss35 | 0.210 | vamp2 | -0.079 |
| atp1b4 | 0.207 | elavl4 | -0.079 |
| tes | 0.205 | si:dkey-276j7.1 | -0.079 |
| col4a6 | 0.205 | sncb | -0.078 |
| pbxip1b | 0.201 | ywhah | -0.078 |
| clstn2 | 0.200 | stx1b | -0.078 |
| pleca | 0.198 | tmsb | -0.076 |
| hepacama | 0.195 | fscn1a | -0.076 |
| vwa7 | 0.194 | rtn1b | -0.075 |
| zgc:165461 | 0.194 | zc4h2 | -0.074 |
| itgb4 | 0.193 | gng2 | -0.074 |
| sema3ga | 0.192 | snap25a | -0.074 |
| igfbp7 | 0.185 | marcksb | -0.074 |
| slc6a11b | 0.185 | LOC100537384 | -0.071 |
| mdkb | 0.181 | calm1a | -0.071 |
| mdka | 0.175 | tmem59l | -0.070 |
| tgm1l1 | 0.173 | cirbpb | -0.070 |
| crabp1b | 0.168 | h3f3a | -0.070 |
| slc3a2a | 0.165 | tubb2b | -0.069 |
| pald1b | 0.162 | onecut2 | -0.068 |
| dsg2.1 | 0.162 | stmn2a | -0.068 |
| cx43 | 0.161 | myt1a | -0.068 |
| fam181b | 0.160 | gap43 | -0.067 |
| hspb15 | 0.159 | onecut1 | -0.067 |