pre-B-cell leukemia homeobox interacting protein 1b
ZFIN 
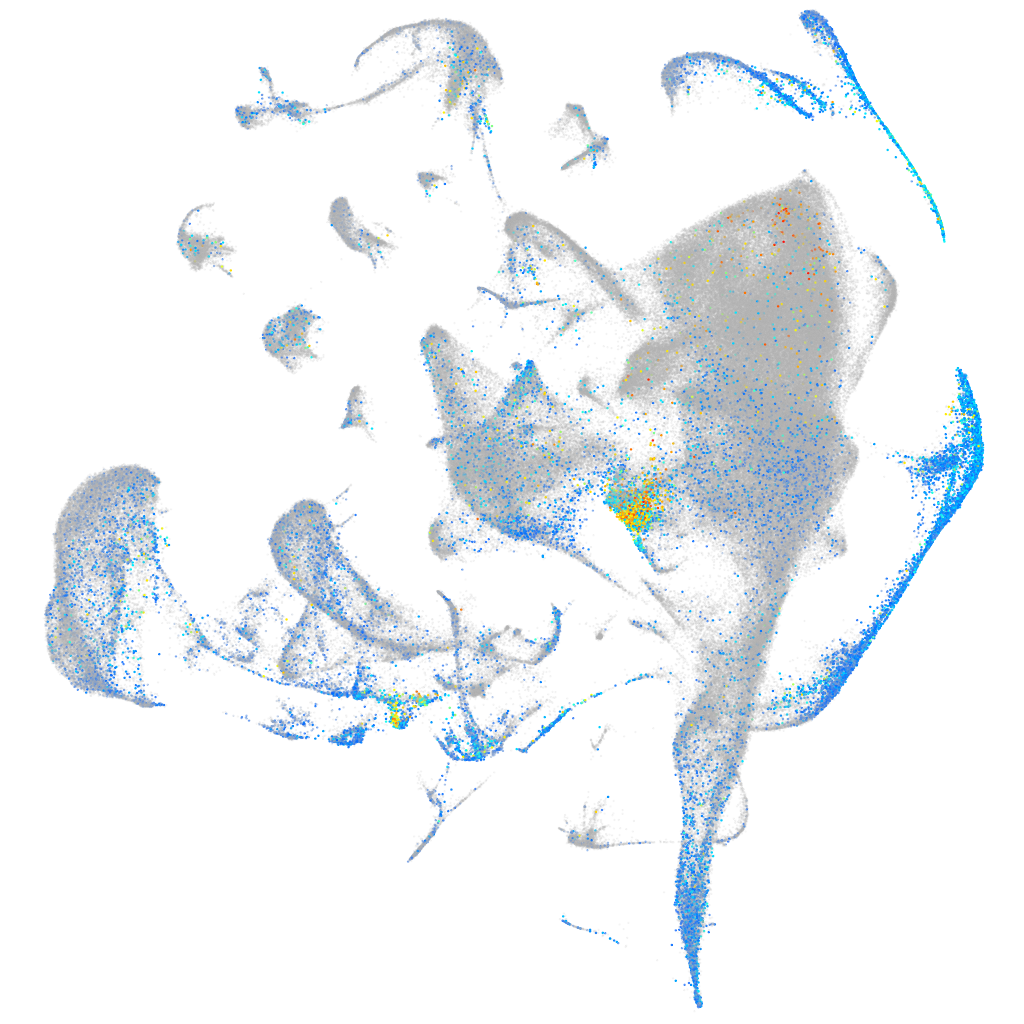
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| hsp90aa1.1 | 0.356 | elavl3 | -0.109 |
| si:ch211-131k2.3 | 0.333 | hmgb1b | -0.108 |
| tnni2b.1 | 0.324 | tuba1c | -0.108 |
| zgc:92429 | 0.315 | tubb5 | -0.102 |
| mss51 | 0.314 | stmn1b | -0.101 |
| unc45b | 0.313 | gpm6ab | -0.100 |
| zgc:92518 | 0.310 | nova2 | -0.099 |
| myog | 0.307 | si:dkey-276j7.1 | -0.097 |
| CR381686.5 | 0.306 | chd4a | -0.095 |
| flncb | 0.304 | zc4h2 | -0.093 |
| smyd1b | 0.304 | tuba1a | -0.089 |
| txlnba | 0.303 | atp6v0cb | -0.087 |
| fbxl22 | 0.302 | epb41a | -0.086 |
| hjv | 0.300 | rtn1a | -0.086 |
| tmx2a | 0.296 | marcksl1a | -0.086 |
| chrng | 0.288 | si:ch211-137a8.4 | -0.085 |
| actc1a | 0.287 | tmeff1b | -0.083 |
| klhl41b | 0.286 | rtn1b | -0.081 |
| tnnt2c | 0.286 | gng3 | -0.080 |
| rbfox1l | 0.283 | hmgb3a | -0.080 |
| trim109 | 0.283 | sncb | -0.080 |
| txlnbb | 0.282 | elavl4 | -0.078 |
| klhl40b | 0.271 | myt1a | -0.076 |
| desma | 0.266 | myt1b | -0.076 |
| myod1 | 0.266 | gng2 | -0.073 |
| asb12b | 0.263 | si:ch211-288g17.3 | -0.073 |
| acta1a | 0.261 | gpm6aa | -0.072 |
| chrnd | 0.260 | stx1b | -0.072 |
| mir133c | 0.254 | FO082781.1 | -0.071 |
| obsl1a | 0.251 | marcksb | -0.071 |
| zgc:101853 | 0.251 | nkain1 | -0.071 |
| cldn7a | 0.250 | rnasekb | -0.071 |
| BX276101.1 | 0.247 | atpv0e2 | -0.070 |
| rbm24a | 0.242 | gap43 | -0.069 |
| mymk | 0.241 | si:ch73-386h18.1 | -0.069 |