kelch-like family member 40b
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line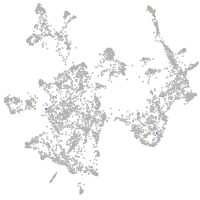 |
epidermis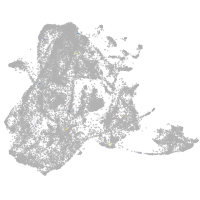 |
periderm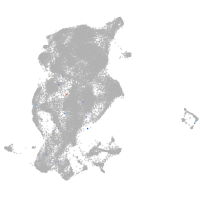 |
fin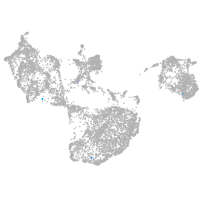 |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| tnni2b.1 | 0.593 | hmgb1b | -0.087 |
| txlnba | 0.587 | marcksl1a | -0.080 |
| si:ch211-131k2.3 | 0.587 | si:ch211-288g17.3 | -0.075 |
| tmx2a | 0.586 | chd4a | -0.071 |
| txlnbb | 0.585 | cbx5 | -0.070 |
| CR381686.5 | 0.564 | stmn1a | -0.070 |
| smyd1b | 0.564 | pcna | -0.065 |
| zgc:92518 | 0.556 | rbbp4 | -0.063 |
| tnnt2c | 0.555 | tuba1c | -0.063 |
| mss51 | 0.554 | seta | -0.062 |
| hjv | 0.553 | ywhaqb | -0.062 |
| rbfox1l | 0.537 | nova2 | -0.061 |
| srl | 0.531 | pfn2 | -0.061 |
| desma | 0.526 | si:ch211-137a8.4 | -0.061 |
| hsp90aa1.1 | 0.524 | ctsla | -0.060 |
| actc1a | 0.524 | mcm7 | -0.059 |
| asb12b | 0.520 | tuba1a | -0.059 |
| chrng | 0.519 | hmgn3 | -0.058 |
| klhl41b | 0.515 | mdkb | -0.058 |
| fbxl22 | 0.514 | morf4l1 | -0.058 |
| acta1a | 0.507 | anp32a | -0.057 |
| obsl1a | 0.505 | gapdhs | -0.057 |
| chrnd | 0.504 | mif | -0.057 |
| klhl31 | 0.504 | prdx2 | -0.057 |
| zgc:101853 | 0.499 | gpm6aa | -0.056 |
| unc45b | 0.499 | h2afva | -0.056 |
| CABZ01072309.2 | 0.498 | lasp1 | -0.056 |
| cacng1a | 0.494 | ywhaqa | -0.056 |
| dhrs7ca | 0.492 | zc4h2 | -0.056 |
| zgc:92429 | 0.484 | atrx | -0.054 |
| filip1a | 0.476 | si:ch211-156b7.4 | -0.054 |
| fitm1 | 0.471 | ckbb | -0.053 |
| cav3 | 0.467 | dut | -0.053 |
| tmem38a | 0.464 | foxp4 | -0.053 |
| alpk3a | 0.461 | tuba8l | -0.053 |




