retinal homeobox gene 3
ZFIN 
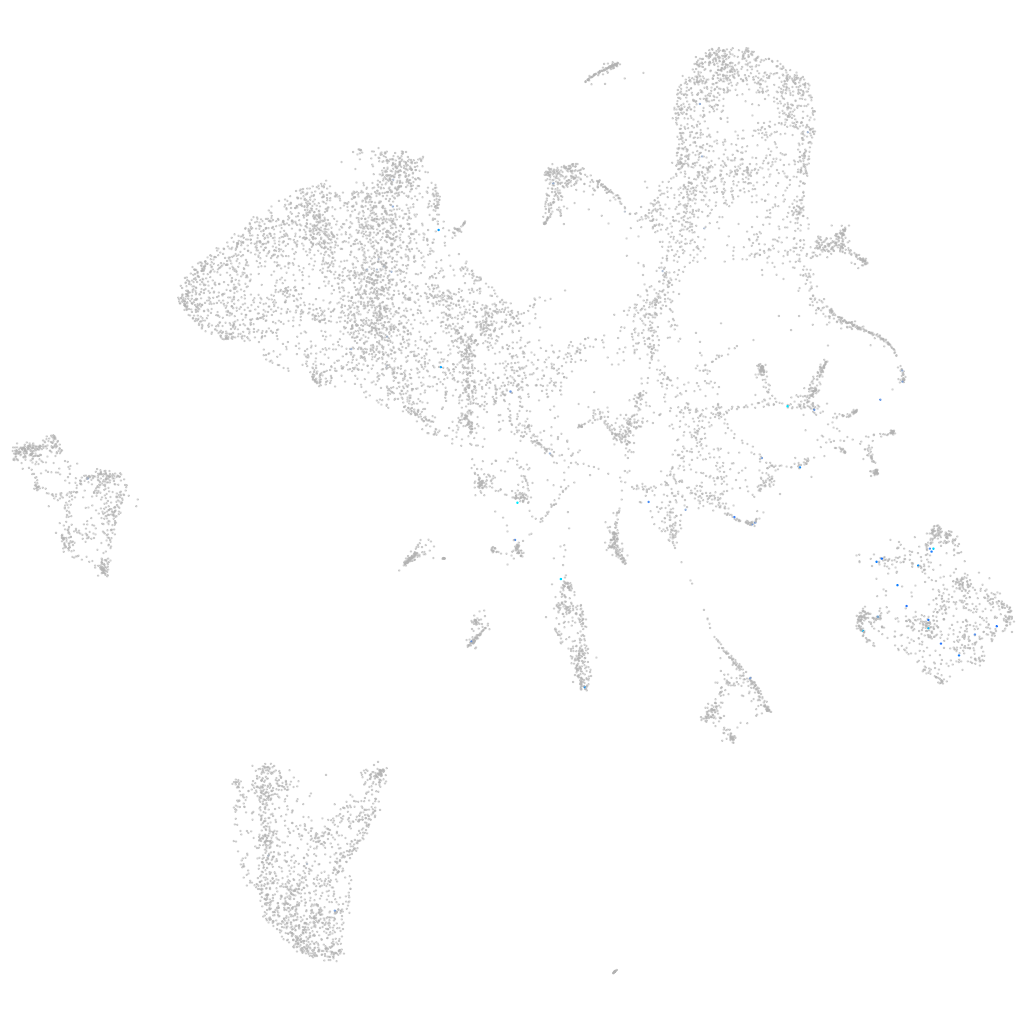
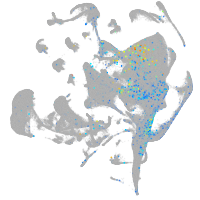
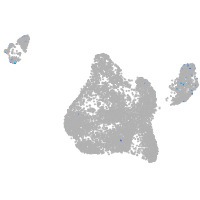
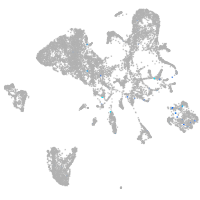
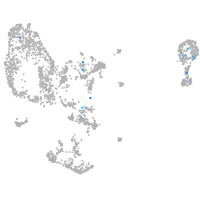
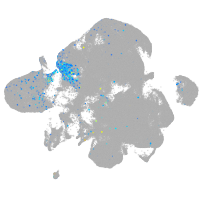
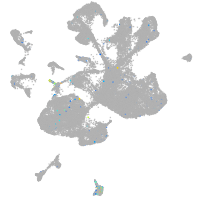
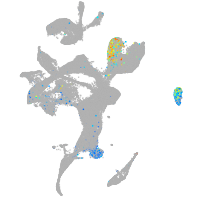
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| rrh | 0.279 | rpl37 | -0.066 |
| BX005056.1 | 0.260 | zgc:114188 | -0.065 |
| six3b | 0.254 | rps17 | -0.063 |
| lcp1 | 0.249 | rps10 | -0.063 |
| zgc:175177 | 0.235 | nme2b.1 | -0.057 |
| CABZ01053748.1 | 0.235 | ahcy | -0.056 |
| LOC110438375 | 0.222 | eef1da | -0.053 |
| CABZ01077555.1 | 0.220 | prdx2 | -0.049 |
| CR354610.2 | 0.219 | rpl17 | -0.049 |
| slc1a7a | 0.211 | gapdh | -0.047 |
| pomca | 0.207 | zgc:158463 | -0.047 |
| CU929418.2 | 0.199 | adh5 | -0.047 |
| BX649622.1 | 0.197 | sdhdb | -0.047 |
| LOC108190098 | 0.197 | atp5mc1 | -0.045 |
| ca14 | 0.191 | atp5pf | -0.045 |
| LOC108183629 | 0.191 | cox6b1 | -0.045 |
| si:ch211-207l14.1 | 0.188 | pklr | -0.044 |
| esamb | 0.183 | krtcap2 | -0.044 |
| hyal6 | 0.183 | gsta.1 | -0.044 |
| LOC110438742 | 0.180 | nupr1b | -0.044 |
| zgc:165604 | 0.169 | eef1b2 | -0.044 |
| serinc4 | 0.168 | suclg1 | -0.044 |
| arid5a | 0.164 | gamt | -0.043 |
| XLOC-026713 | 0.160 | prdx4 | -0.043 |
| crabp1a | 0.160 | gstt1a | -0.043 |
| six6b | 0.159 | cox5ab | -0.041 |
| cspp1a | 0.158 | glud1b | -0.041 |
| LOC101883622 | 0.156 | sod1 | -0.041 |
| plxnb2b | 0.156 | COX7A2 (1 of many) | -0.041 |
| stk17a | 0.156 | gstp1 | -0.041 |
| fgf8b | 0.155 | atp5f1b | -0.041 |
| CT573366.1 | 0.153 | rpl9 | -0.041 |
| LOC101885011 | 0.152 | hadh | -0.041 |
| mkrn2os.2 | 0.148 | fbp1b | -0.040 |
| AL953841.2 | 0.147 | minos1 | -0.040 |