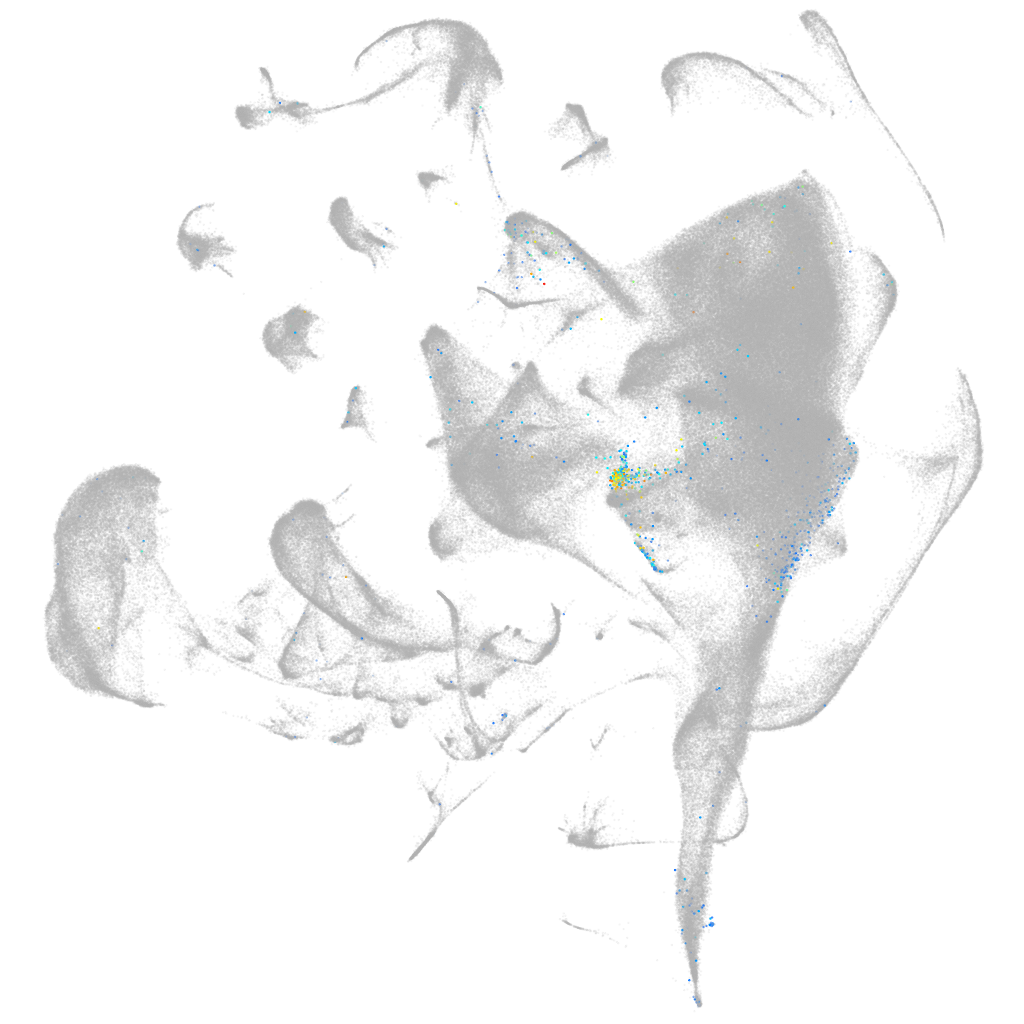
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| ca14 | 0.346 | si:dkey-56m19.5 | -0.020 |
| hyal6 | 0.335 | elavl3 | -0.018 |
| zgc:165604 | 0.247 | tmsb | -0.018 |
| rlbp1a | 0.239 | cfl1l | -0.017 |
| CU929418.2 | 0.222 | cyt1l | -0.017 |
| ush1ga | 0.182 | krt4 | -0.017 |
| slc2a1a | 0.165 | nova1 | -0.017 |
| gpr37a | 0.163 | tubb5 | -0.017 |
| aqp4 | 0.145 | aep1 | -0.016 |
| calml4b | 0.145 | hmgb3a | -0.016 |
| LOC101886429 | 0.141 | wu:fb18f06 | -0.016 |
| inhbab | 0.137 | arhgdig | -0.015 |
| LO018231.1 | 0.121 | cyt1 | -0.015 |
| ush1c | 0.120 | elavl4 | -0.015 |
| mlc1 | 0.119 | epcam | -0.015 |
| fzd5 | 0.118 | stmn1b | -0.015 |
| acbd7 | 0.117 | tp53inp2 | -0.015 |
| efhd1 | 0.116 | agr1 | -0.014 |
| abi3a | 0.115 | anxa1a | -0.014 |
| crabp1a | 0.115 | anxa1c | -0.014 |
| sntb1 | 0.111 | chd4a | -0.014 |
| glula | 0.110 | gap43 | -0.014 |
| ppp1r14aa | 0.110 | gsnb | -0.014 |
| cavin2a | 0.108 | icn2 | -0.014 |
| lmo7b | 0.108 | lye | -0.014 |
| syngr2b | 0.108 | myt1b | -0.014 |
| zgc:109949 | 0.107 | pycard | -0.014 |
| mt2 | 0.104 | rbfox2 | -0.014 |
| rergla | 0.101 | s100v1 | -0.014 |
| vsx2 | 0.101 | sox11b | -0.014 |
| fgf24 | 0.097 | stmn2a | -0.014 |
| tgfb3 | 0.097 | tmsb1 | -0.014 |
| cdo1 | 0.094 | zfhx3 | -0.014 |
| cldn5b | 0.093 | zgc:153284 | -0.014 |
| col15a1b | 0.093 | zgc:193505 | -0.014 |