RHO family interacting cell polarization regulator 2
ZFIN 
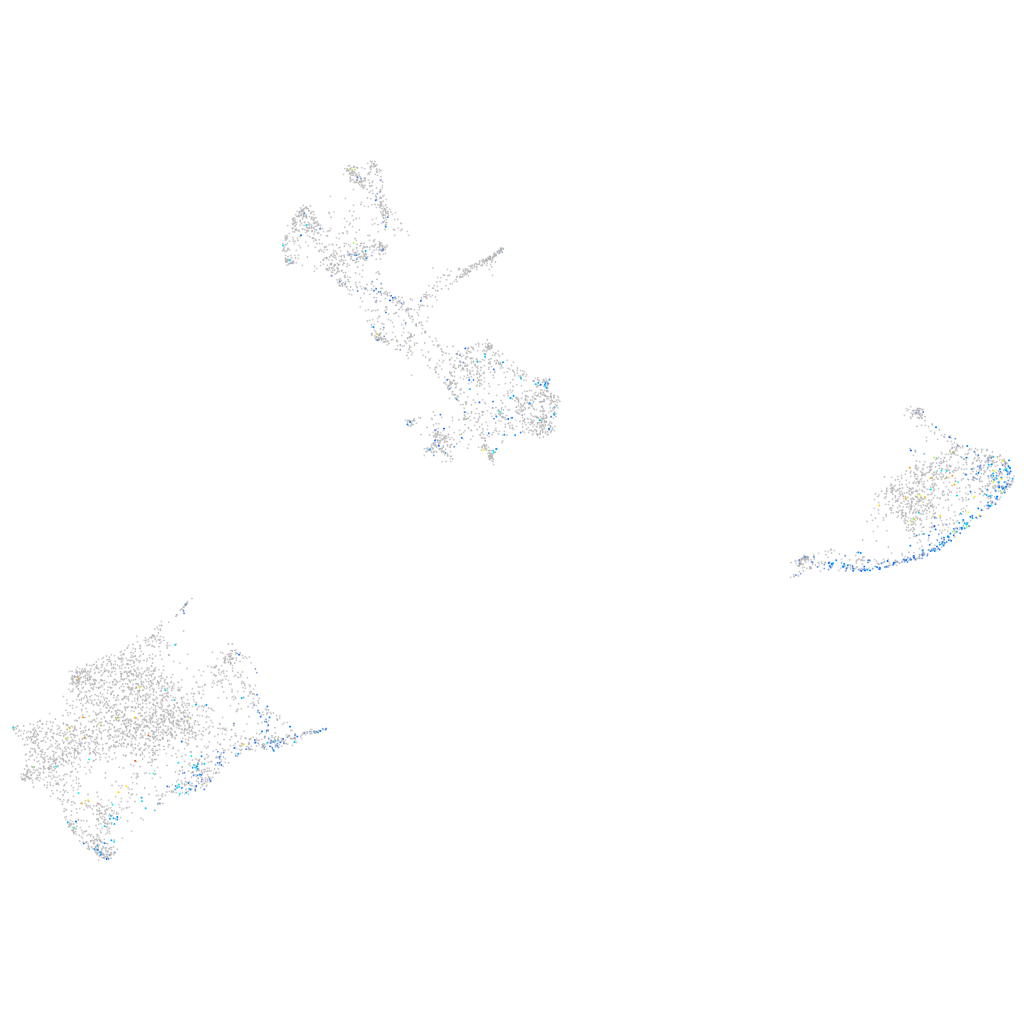
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| SPAG9 | 0.239 | paics | -0.094 |
| kita | 0.216 | uraha | -0.091 |
| lamp1a | 0.192 | CABZ01021592.1 | -0.066 |
| tyr | 0.192 | defbl1 | -0.065 |
| oca2 | 0.183 | apoda.1 | -0.063 |
| slc39a10 | 0.174 | mdh1aa | -0.061 |
| pmela | 0.173 | si:ch211-256m1.8 | -0.060 |
| slc24a5 | 0.172 | si:dkey-251i10.2 | -0.059 |
| slc7a5 | 0.169 | pvalb1 | -0.058 |
| dct | 0.168 | lypc | -0.056 |
| tyrp1a | 0.167 | rps5 | -0.055 |
| CDK18 | 0.167 | gpnmb | -0.054 |
| tyrp1b | 0.165 | rpl19 | -0.053 |
| prkar1b | 0.160 | si:dkey-197i20.6 | -0.053 |
| mlpha | 0.158 | rps23 | -0.052 |
| zgc:91968 | 0.158 | phyhd1 | -0.051 |
| pknox1.2 | 0.158 | rplp1 | -0.051 |
| tfap2e | 0.158 | atic | -0.051 |
| slc37a2 | 0.157 | rps8a | -0.051 |
| bace2 | 0.155 | krt18b | -0.051 |
| mitfa | 0.153 | alx4a | -0.050 |
| tfap2a | 0.153 | si:dkey-151g10.6 | -0.050 |
| slc29a3 | 0.152 | hbae3 | -0.050 |
| kcnj13 | 0.152 | unm-sa821 | -0.048 |
| lyst | 0.150 | tagln3b | -0.047 |
| atp6v0ca | 0.149 | hbbe1.3 | -0.047 |
| AL935199.1 | 0.148 | prdx5 | -0.046 |
| mchr2 | 0.148 | prdx1 | -0.045 |
| zdhhc2 | 0.147 | si:ch211-105c13.3 | -0.043 |
| pcdh10a | 0.147 | alx1 | -0.043 |
| atp6v0a2b | 0.146 | alx4b | -0.043 |
| slc6a15 | 0.146 | sytl2b | -0.042 |
| slc24a4a | 0.145 | cyb5a | -0.042 |
| map7a | 0.144 | prps1a | -0.042 |
| hsd20b2 | 0.144 | rplp0 | -0.042 |