RIB43A domain with coiled-coils 2
ZFIN 
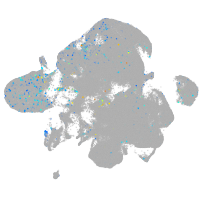
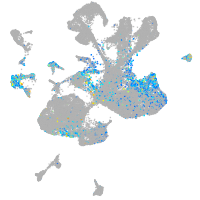
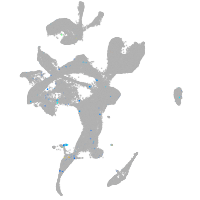
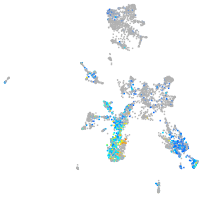
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| daw1 | 0.205 | ptmaa | -0.042 |
| ccdc173 | 0.163 | elavl3 | -0.038 |
| dydc2 | 0.154 | COX3 | -0.036 |
| nme5 | 0.153 | tmsb4x | -0.035 |
| ccdc114 | 0.152 | gpm6ab | -0.035 |
| si:ch211-155m12.5 | 0.151 | stmn1b | -0.033 |
| foxj1a | 0.140 | fabp3 | -0.033 |
| pih1d3 | 0.136 | FO082781.1 | -0.033 |
| cfap126 | 0.136 | gpm6aa | -0.033 |
| rsph4a | 0.134 | elavl4 | -0.032 |
| rsph9 | 0.131 | mt-atp6 | -0.032 |
| pacrg | 0.130 | ckbb | -0.031 |
| enkur | 0.130 | rpl37 | -0.031 |
| spag8 | 0.129 | tuba1c | -0.030 |
| ropn1l | 0.129 | h3f3a | -0.030 |
| ccdc151 | 0.128 | mt-co2 | -0.030 |
| cfap52 | 0.126 | tmsb | -0.029 |
| si:ch211-163l21.10 | 0.125 | mab21l1 | -0.029 |
| si:ch211-248e11.2 | 0.124 | zgc:114188 | -0.028 |
| CRACR2A | 0.120 | mt-nd1 | -0.027 |
| si:ch211-163l21.7 | 0.116 | pvalb1 | -0.026 |
| prdx1 | 0.116 | zgc:158463 | -0.026 |
| dnaaf1 | 0.115 | mab21l2 | -0.026 |
| catip | 0.111 | nova2 | -0.026 |
| dnaaf4 | 0.110 | bach2b | -0.026 |
| dnai1.2 | 0.109 | ppiab | -0.025 |
| tbata | 0.109 | rbfox1 | -0.025 |
| cfap53 | 0.109 | cadm3 | -0.025 |
| morn3 | 0.109 | actc1b | -0.024 |
| si:dkey-27p23.3 | 0.108 | tubb5 | -0.024 |
| zc2hc1c | 0.108 | pvalb2 | -0.024 |
| meig1 | 0.107 | tubb2b | -0.024 |
| spag6 | 0.107 | myt1la | -0.024 |
| armc3 | 0.107 | rps10 | -0.024 |
| zmynd10 | 0.107 | tnrc6c1 | -0.024 |