RNA binding motif protein 39b
ZFIN 
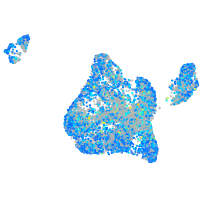
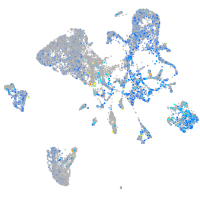
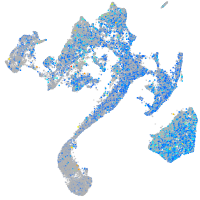
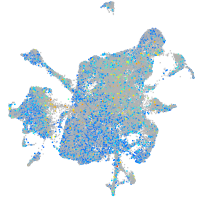
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| nop58 | 0.059 | crybb1l2 | -0.029 |
| nop56 | 0.058 | crybb1l1 | -0.028 |
| ncl | 0.058 | crygn2 | -0.028 |
| npm1a | 0.056 | cryba1b | -0.028 |
| wdr43 | 0.055 | mipb | -0.027 |
| snu13b | 0.055 | cryba2a | -0.027 |
| dkc1 | 0.055 | COX3 | -0.026 |
| ebna1bp2 | 0.054 | mipa | -0.026 |
| rpl7l1 | 0.052 | cryba4 | -0.026 |
| hnrnpabb | 0.052 | lim2.4 | -0.026 |
| nhp2 | 0.051 | cryba2b | -0.026 |
| mak16 | 0.051 | crybb1 | -0.026 |
| pprc1 | 0.051 | crygmx | -0.026 |
| hnrnpaba | 0.051 | cryba1l2 | -0.026 |
| syncrip | 0.051 | cryba1l1 | -0.026 |
| apoeb | 0.051 | BX545855.1 | -0.025 |
| ddx5 | 0.051 | capn3a | -0.025 |
| ppan | 0.051 | crygm2d8 | -0.024 |
| aldoaa | 0.050 | crygm2d13 | -0.024 |
| hnrnpa0b | 0.050 | grifin | -0.024 |
| fbl | 0.050 | cryaa | -0.024 |
| srsf3b | 0.050 | lim2.3 | -0.024 |
| nop2 | 0.050 | crygm2d10 | -0.024 |
| nop10 | 0.049 | crybb1l3 | -0.024 |
| sumo3a | 0.049 | lim2.5 | -0.023 |
| ilf2 | 0.049 | b3gnt5a | -0.023 |
| gar1 | 0.049 | endou2 | -0.023 |
| setb | 0.049 | si:ch211-255g12.6 | -0.022 |
| si:ch211-51e12.7 | 0.048 | rpl37 | -0.022 |
| prmt1 | 0.048 | crygm2d12 | -0.022 |
| hspb1 | 0.048 | lim2.1 | -0.021 |
| lyar | 0.048 | tubb6 | -0.021 |
| eif5 | 0.048 | crygm2d1 | -0.021 |
| hmgn7 | 0.048 | si:ch211-214j24.14 | -0.021 |
| magoh | 0.048 | crygm2d20 | -0.021 |