"retinoic acid receptor, alpha b"
ZFIN 
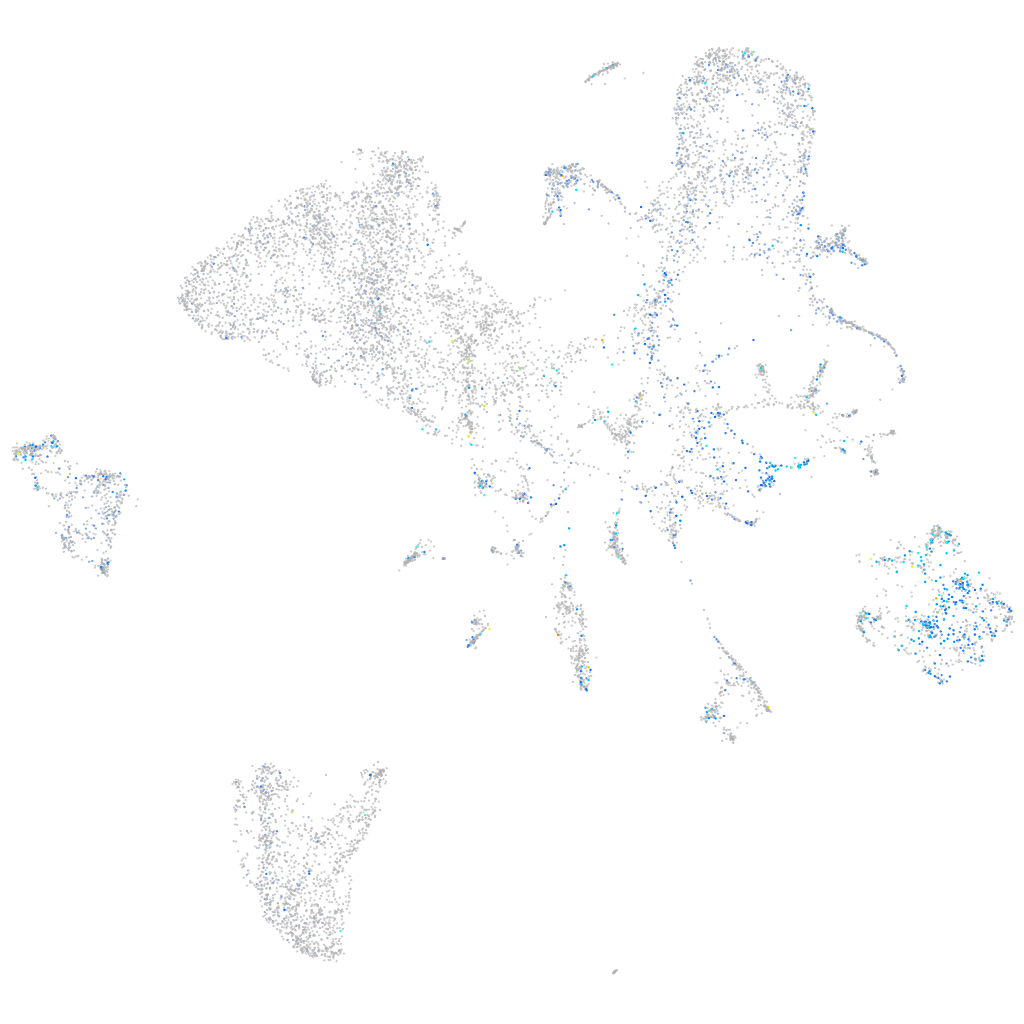
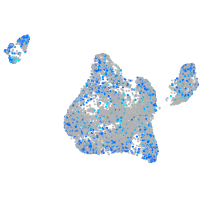
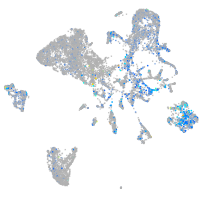
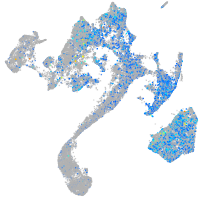
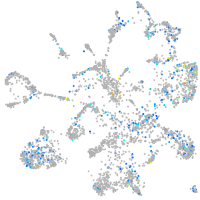
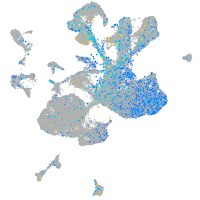
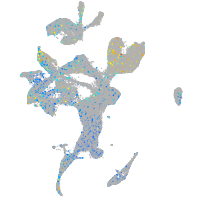
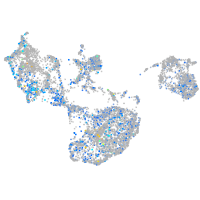
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| hspb1 | 0.265 | gamt | -0.196 |
| cx43.4 | 0.260 | bhmt | -0.192 |
| hnrnpub | 0.259 | ahcy | -0.186 |
| cdx4 | 0.259 | gapdh | -0.178 |
| nucks1a | 0.252 | gatm | -0.178 |
| hnrnpm | 0.244 | agxtb | -0.175 |
| akap12b | 0.242 | apoa1b | -0.171 |
| khdrbs1a | 0.240 | mat1a | -0.170 |
| syncrip | 0.240 | aqp12 | -0.169 |
| ilf3b | 0.236 | apoa2 | -0.169 |
| hmga1a | 0.236 | nupr1b | -0.166 |
| marcksb | 0.235 | rpl37 | -0.166 |
| acin1a | 0.235 | zgc:92744 | -0.164 |
| si:ch73-1a9.3 | 0.233 | rps10 | -0.162 |
| hnrnpa1b | 0.231 | grhprb | -0.161 |
| si:ch73-281n10.2 | 0.231 | agxta | -0.157 |
| marcksl1b | 0.229 | pnp4b | -0.155 |
| qkia | 0.228 | rbp2b | -0.155 |
| smarca4a | 0.227 | fabp10a | -0.155 |
| seta | 0.227 | kng1 | -0.154 |
| asph | 0.226 | fabp3 | -0.154 |
| hnrnpa1a | 0.226 | zgc:123103 | -0.153 |
| snrnp70 | 0.226 | eno3 | -0.153 |
| zgc:110425 | 0.226 | pgm1 | -0.152 |
| smc1al | 0.226 | serpina1l | -0.152 |
| hnrnpaba | 0.225 | hpda | -0.151 |
| cdh6 | 0.222 | ttc36 | -0.151 |
| hmgn6 | 0.222 | apom | -0.151 |
| sp5l | 0.221 | zgc:114188 | -0.150 |
| brd3a | 0.221 | rbp4 | -0.150 |
| ube2e1 | 0.220 | fgg | -0.149 |
| safb | 0.220 | ces2 | -0.149 |
| vox | 0.219 | uox | -0.149 |
| apela | 0.219 | serpina1 | -0.149 |
| NC-002333.4 | 0.219 | tfa | -0.149 |