prostaglandin reductase 1
ZFIN 
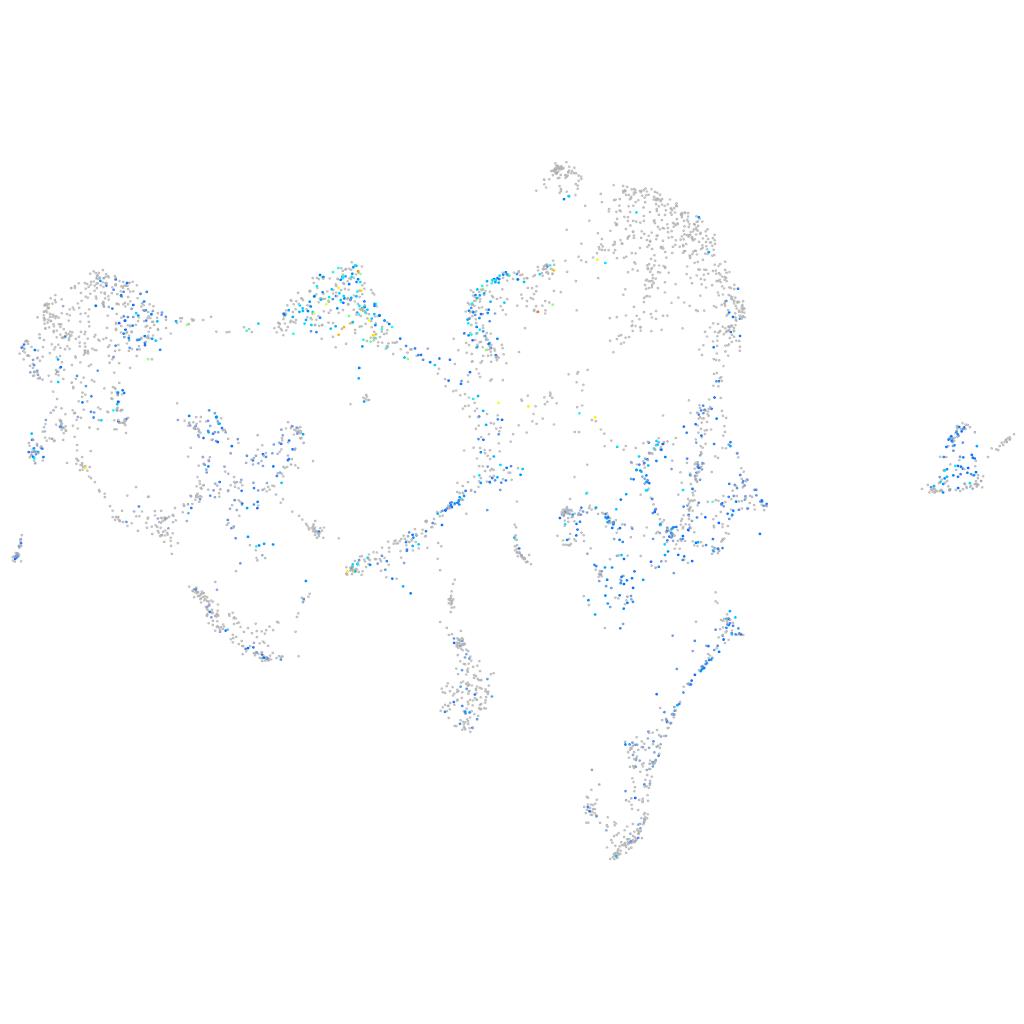
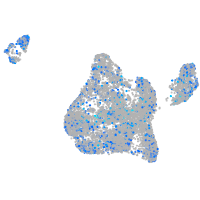
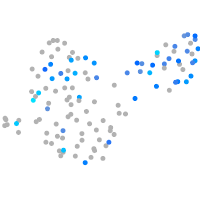
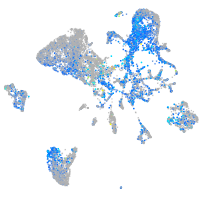
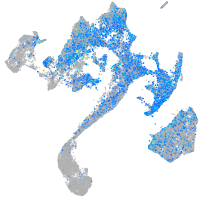
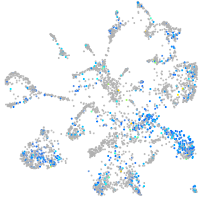
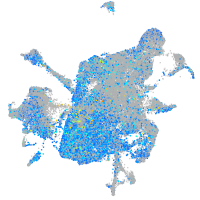
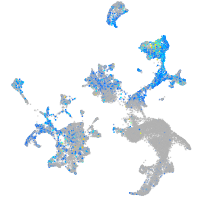
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| si:dkey-11k2.7 | 0.259 | muc5.1 | -0.211 |
| h2afva | 0.238 | b3gnt7 | -0.202 |
| si:dkey-189h5.6 | 0.220 | si:dkey-65b12.6 | -0.201 |
| ddt | 0.220 | agr2 | -0.195 |
| prdx5 | 0.214 | krt5 | -0.180 |
| nudt4a | 0.211 | zgc:92380 | -0.179 |
| elovl1b | 0.210 | atp2a3 | -0.177 |
| ndrg3a | 0.209 | LOC110438378 | -0.170 |
| gsto1 | 0.209 | p2rx1 | -0.167 |
| zgc:110339 | 0.208 | muc5.2 | -0.163 |
| ppil1 | 0.208 | aldob | -0.163 |
| echdc2 | 0.208 | stard10 | -0.162 |
| calm2b | 0.207 | nansb | -0.156 |
| atp1a1a.2 | 0.204 | cmah | -0.147 |
| jagn1b | 0.201 | sytl2b | -0.145 |
| LOC799574 | 0.201 | galnt7 | -0.143 |
| rgs5b | 0.200 | tent5bb | -0.142 |
| atp1a1a.3 | 0.200 | klf3 | -0.142 |
| ncln | 0.198 | si:dkey-33i11.9 | -0.141 |
| clcn2c | 0.198 | calm1b | -0.140 |
| prlra | 0.197 | gng13b | -0.137 |
| trim35-12 | 0.196 | ankrd22 | -0.137 |
| foxi3b | 0.195 | rap1gap | -0.137 |
| arpp19b | 0.194 | chst6 | -0.136 |
| zgc:194246 | 0.192 | zgc:112994 | -0.135 |
| zgc:163083 | 0.192 | galnt12 | -0.134 |
| si:dkey-33i11.4 | 0.192 | pleca | -0.133 |
| slc4a4b | 0.192 | icn | -0.133 |
| atp5if1a | 0.191 | si:dkey-246j7.1 | -0.133 |
| cox8b | 0.190 | tcima | -0.132 |
| atp5fa1 | 0.188 | cers3b | -0.131 |
| atp1b1b | 0.185 | grb10a | -0.131 |
| si:ch211-153b23.5 | 0.185 | si:ch73-306e8.2 | -0.130 |
| atp5mc3b | 0.183 | fer1l6 | -0.130 |
| atp5f1b | 0.183 | SPDEF | -0.129 |