zgc:194246
ZFIN 
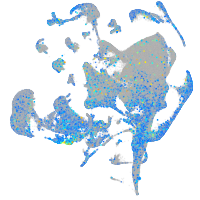
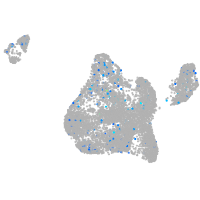
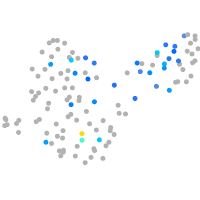
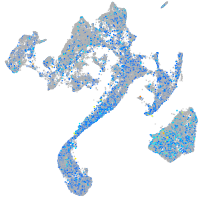
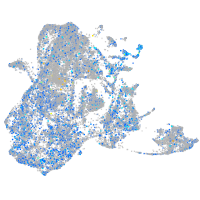
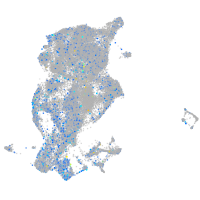
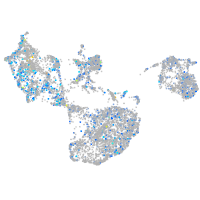
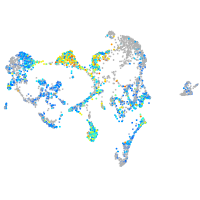
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| foxi3b | 0.236 | gpm6aa | -0.104 |
| si:dkey-33i11.4 | 0.227 | tuba1c | -0.103 |
| atp1a1a.2 | 0.182 | nova2 | -0.097 |
| LOC799574 | 0.175 | elavl3 | -0.092 |
| slc12a10.2 | 0.170 | stmn1b | -0.086 |
| si:dkey-189h5.6 | 0.165 | rtn1a | -0.085 |
| mal2 | 0.163 | gpm6ab | -0.084 |
| trim35-12 | 0.162 | tuba1a | -0.084 |
| si:ch211-270g19.5 | 0.158 | ckbb | -0.079 |
| atp1b1b | 0.155 | epb41a | -0.075 |
| atp1a1a.3 | 0.152 | tubb5 | -0.075 |
| clcn2c | 0.151 | cadm3 | -0.074 |
| cldnh | 0.140 | myt1b | -0.071 |
| BX000438.2 | 0.139 | atp6v0cb | -0.070 |
| si:ch73-359m17.9 | 0.139 | hmgb1b | -0.070 |
| sgk2a | 0.138 | celf2 | -0.069 |
| sstr5 | 0.128 | mdkb | -0.069 |
| foxi3a | 0.127 | fabp7a | -0.068 |
| kcnj1a.1 | 0.125 | si:dkey-276j7.1 | -0.068 |
| nipal4 | 0.123 | si:ch211-137a8.4 | -0.067 |
| si:dkeyp-97e7.9 | 0.123 | myt1a | -0.066 |
| dnase1l4.1 | 0.122 | rnasekb | -0.065 |
| prom2 | 0.121 | cspg5a | -0.064 |
| prg4a | 0.120 | scrt2 | -0.064 |
| mat2al | 0.119 | gng3 | -0.063 |
| echdc2 | 0.116 | tmsb | -0.063 |
| si:ch73-31d8.2 | 0.115 | chd4a | -0.061 |
| kcnj1a.2 | 0.115 | CU467822.1 | -0.061 |
| hepacam2 | 0.114 | CU634008.1 | -0.061 |
| bik | 0.112 | hmgb3a | -0.060 |
| dmrt2a | 0.112 | sncb | -0.060 |
| rgs5b | 0.112 | FO082781.1 | -0.059 |
| mhc2dbb | 0.109 | ncam1a | -0.059 |
| rnaseka | 0.107 | marcksl1a | -0.059 |
| slc4a4b | 0.107 | fam168a | -0.058 |