growth factor receptor-bound protein 10a
ZFIN 
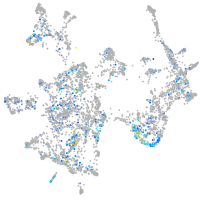
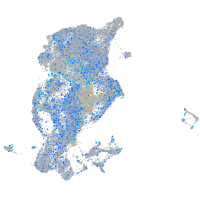
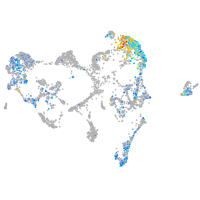
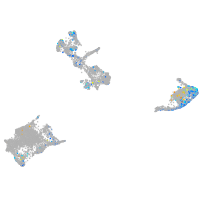
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| si:ch211-145b13.6 | 0.201 | gpm6aa | -0.105 |
| plvapb | 0.199 | tuba1c | -0.104 |
| clec14a | 0.193 | stmn1b | -0.097 |
| si:dkeyp-97a10.2 | 0.193 | elavl3 | -0.095 |
| myct1a | 0.189 | rtn1a | -0.092 |
| kdr | 0.187 | hmgb1b | -0.090 |
| kdrl | 0.187 | ckbb | -0.087 |
| cdh5 | 0.186 | hmgb3a | -0.085 |
| pecam1 | 0.182 | tmsb | -0.083 |
| tie1 | 0.182 | si:dkey-276j7.1 | -0.082 |
| exoc3l2a | 0.181 | fabp3 | -0.081 |
| flt1 | 0.173 | tuba1a | -0.081 |
| ecscr | 0.167 | tubb5 | -0.079 |
| she | 0.166 | chd4a | -0.077 |
| si:ch73-334d15.2 | 0.166 | gng3 | -0.077 |
| etv2 | 0.163 | zc4h2 | -0.077 |
| tmem88a | 0.161 | marcksb | -0.076 |
| si:ch211-33e4.2 | 0.159 | nova2 | -0.076 |
| egfl7 | 0.151 | atp6v0cb | -0.073 |
| cavin2b | 0.150 | celf2 | -0.072 |
| notchl | 0.148 | gpm6ab | -0.071 |
| pcdh12 | 0.148 | myt1b | -0.071 |
| ahnak | 0.146 | sncb | -0.071 |
| krt8 | 0.146 | cspg5a | -0.070 |
| rasip1 | 0.146 | epb41a | -0.070 |
| slc9a3r1b | 0.145 | mdkb | -0.070 |
| igfbp7 | 0.144 | myt1a | -0.069 |
| LOC101882770 | 0.144 | si:ch211-137a8.4 | -0.069 |
| myct1b | 0.144 | si:ch211-288g17.3 | -0.069 |
| plk2b | 0.144 | fez1 | -0.068 |
| srgn | 0.144 | cadm3 | -0.067 |
| fgd5a | 0.143 | rtn1b | -0.067 |
| gpr182 | 0.143 | sox11b | -0.067 |
| ramp2 | 0.143 | rnasekb | -0.066 |
| si:ch211-248e11.2 | 0.143 | scrt2 | -0.066 |