peroxiredoxin 6
ZFIN 
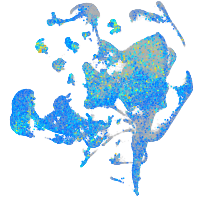
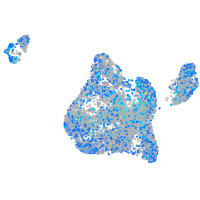
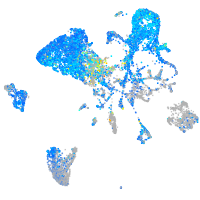
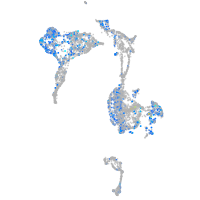
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| fabp7a | 0.199 | elavl3 | -0.143 |
| CU467822.1 | 0.156 | myt1b | -0.114 |
| atp1b4 | 0.153 | onecut1 | -0.107 |
| atp1a1b | 0.152 | myt1a | -0.100 |
| psat1 | 0.150 | lin28a | -0.099 |
| GCA | 0.143 | stxbp1a | -0.098 |
| sod1 | 0.142 | onecut2 | -0.098 |
| eef1da | 0.136 | snap25a | -0.097 |
| ppap2d | 0.134 | elavl4 | -0.096 |
| prdx2 | 0.133 | pik3r3b | -0.092 |
| zgc:165461 | 0.133 | stx1b | -0.092 |
| slc1a2b | 0.132 | gng2 | -0.091 |
| cd82a | 0.132 | syt2a | -0.090 |
| gnai2a | 0.130 | zc4h2 | -0.089 |
| CR848047.1 | 0.130 | sncb | -0.088 |
| mdka | 0.129 | h3f3d | -0.088 |
| si:dkey-7j14.6 | 0.128 | cplx2 | -0.087 |
| slc1a3b | 0.126 | rbfox1 | -0.085 |
| si:ch211-66e2.5 | 0.126 | CABZ01075068.1 | -0.084 |
| si:ch211-251b21.1 | 0.122 | LOC100537384 | -0.084 |
| gpm6bb | 0.122 | cplx2l | -0.084 |
| sod2 | 0.122 | gng3 | -0.084 |
| si:ch211-156b7.4 | 0.121 | scrt2 | -0.083 |
| mif | 0.121 | stmn1b | -0.083 |
| glula | 0.119 | zfhx3 | -0.082 |
| id1 | 0.119 | zgc:100920 | -0.081 |
| ppa1b | 0.117 | cdkn1ca | -0.081 |
| si:ch1073-303k11.2 | 0.117 | hoxc3a | -0.080 |
| sox2 | 0.117 | vamp2 | -0.080 |
| hmga2 | 0.115 | ptmab | -0.079 |
| actb1 | 0.114 | stmn2a | -0.079 |
| zgc:162944 | 0.114 | tmem59l | -0.078 |
| atp5mc3a | 0.114 | gjd1a | -0.078 |
| slc3a2a | 0.114 | dlb | -0.078 |
| cspg5b | 0.112 | aplp1 | -0.077 |