peroxiredoxin 6
ZFIN 
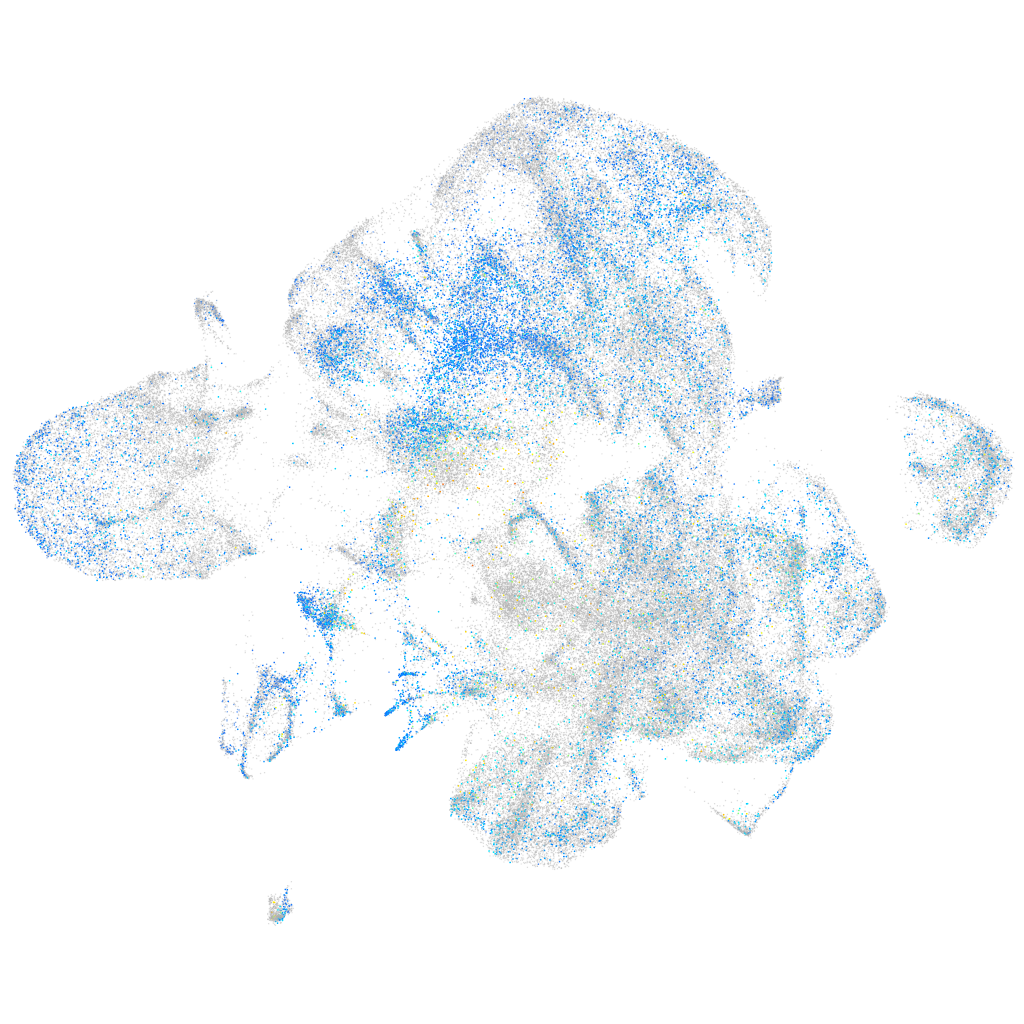
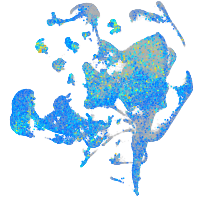
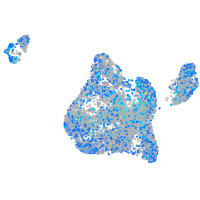
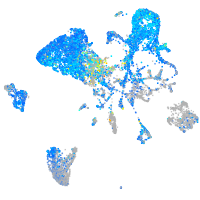
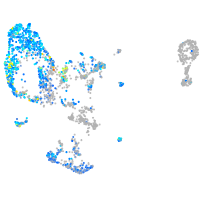
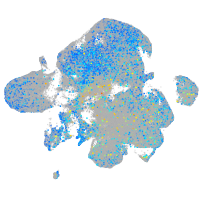
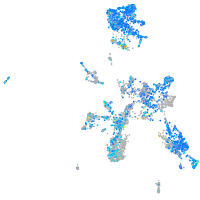
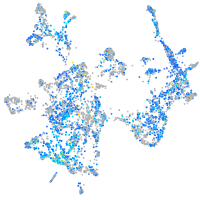
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| ahcy | 0.135 | elavl3 | -0.061 |
| si:ch211-156b7.4 | 0.132 | marcksl1b | -0.060 |
| fabp7a | 0.121 | onecut1 | -0.056 |
| eef1da | 0.121 | stx1b | -0.054 |
| ppa1b | 0.112 | rtn1a | -0.054 |
| stmn1a | 0.111 | atp6v0cb | -0.052 |
| prdx2 | 0.108 | gpm6aa | -0.052 |
| nme2b.1 | 0.105 | NC-002333.4 | -0.051 |
| sod1 | 0.103 | CR383676.1 | -0.051 |
| tuba8l | 0.100 | snap25a | -0.051 |
| dut | 0.097 | sncb | -0.050 |
| adh5 | 0.096 | nr6a1a | -0.050 |
| zgc:56493 | 0.095 | CABZ01075068.1 | -0.049 |
| mif | 0.094 | gpm6ab | -0.049 |
| zgc:162944 | 0.094 | lin28a | -0.048 |
| aldh7a1 | 0.093 | hspb1 | -0.048 |
| psat1 | 0.092 | stxbp1a | -0.047 |
| eef1b2 | 0.092 | LOC100537384 | -0.047 |
| hspe1 | 0.088 | aplp1 | -0.047 |
| atp5mc3b | 0.088 | syt2a | -0.046 |
| atp5mc1 | 0.087 | elavl4 | -0.045 |
| gcshb | 0.087 | gabrb4 | -0.045 |
| atp5mc3a | 0.087 | nrxn1a | -0.045 |
| chchd10 | 0.086 | BX927258.1 | -0.045 |
| nutf2l | 0.085 | vamp2 | -0.045 |
| pa2g4b | 0.085 | sv2a | -0.044 |
| rps17 | 0.085 | zgc:100920 | -0.044 |
| tuba8l4 | 0.083 | atp1a3a | -0.044 |
| sod2 | 0.082 | cplx2 | -0.043 |
| tktb | 0.082 | onecut2 | -0.043 |
| rps2 | 0.081 | syt1a | -0.043 |
| rpl21 | 0.081 | gpr85 | -0.042 |
| rps8a | 0.081 | stmn1b | -0.042 |
| pcna | 0.080 | rtn1b | -0.042 |
| rps3a | 0.080 | id4 | -0.041 |