PDZ domain containing 1
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros 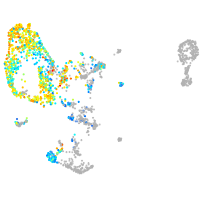 |
neural |
spinal cord / glia |
eye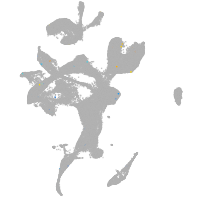 |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous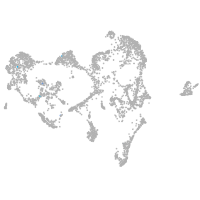 |
pigment cells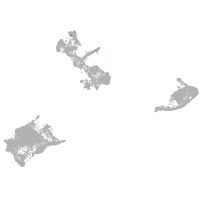 |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| il17d | 0.152 | fabp3 | -0.045 |
| CABZ01032906.1 | 0.121 | vwde | -0.034 |
| pcdh8 | 0.109 | sparc | -0.033 |
| si:dkeyp-77c4.1 | 0.100 | pabpc4 | -0.031 |
| tbx6 | 0.096 | actc1b | -0.030 |
| CABZ01074470.1 | 0.095 | col1a2 | -0.029 |
| draxin | 0.093 | col1a1a | -0.029 |
| ripply2 | 0.092 | marcksl1b | -0.028 |
| CR792438.1 | 0.088 | nid1a | -0.028 |
| CR388165.2 | 0.085 | plecb | -0.028 |
| her11 | 0.084 | alcama | -0.027 |
| CT574585.2 | 0.083 | si:ch211-43f4.1 | -0.027 |
| LOC108191976 | 0.083 | rbp7b | -0.027 |
| RF00319 | 0.082 | eno1a | -0.027 |
| her1 | 0.081 | ckma | -0.027 |
| myf5 | 0.079 | aldoab | -0.027 |
| ripply1 | 0.079 | tmem38a | -0.027 |
| CU914602.1 | 0.078 | ak1 | -0.027 |
| CR759863.2 | 0.078 | ckmb | -0.026 |
| LOC103910639 | 0.077 | sgcd | -0.026 |
| AL954149.1 | 0.076 | srl | -0.026 |
| casr | 0.071 | gapdh | -0.026 |
| ftr11 | 0.071 | ttn.2 | -0.026 |
| apoc1 | 0.071 | atp2a1 | -0.026 |
| LOC101882312 | 0.070 | CABZ01072309.1 | -0.026 |
| mespab | 0.070 | pmp22a | -0.026 |
| tuba8l2 | 0.069 | ldb3a | -0.026 |
| gnaia | 0.069 | txlnbb | -0.025 |
| znfl1k | 0.069 | acta1b | -0.025 |
| mespba | 0.068 | CABZ01078594.1 | -0.025 |
| tjp2b | 0.068 | myl1 | -0.025 |
| ephb3 | 0.068 | mylpfa | -0.025 |
| fn1b | 0.068 | tnnt3a | -0.025 |
| dlc | 0.067 | casq2 | -0.024 |
| cpa6 | 0.066 | neb | -0.024 |