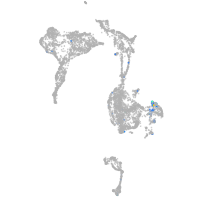
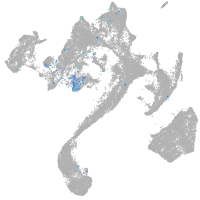
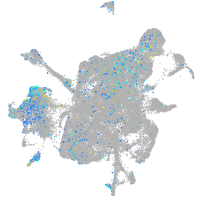
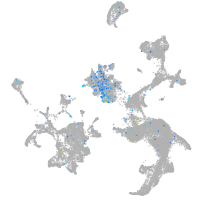
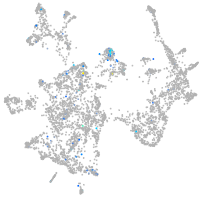
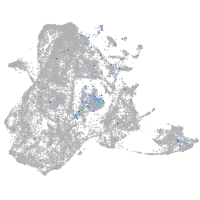
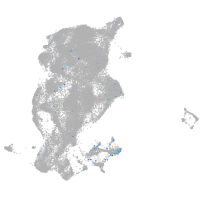
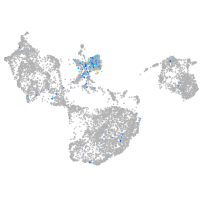
protocadherin 9
ZFIN 
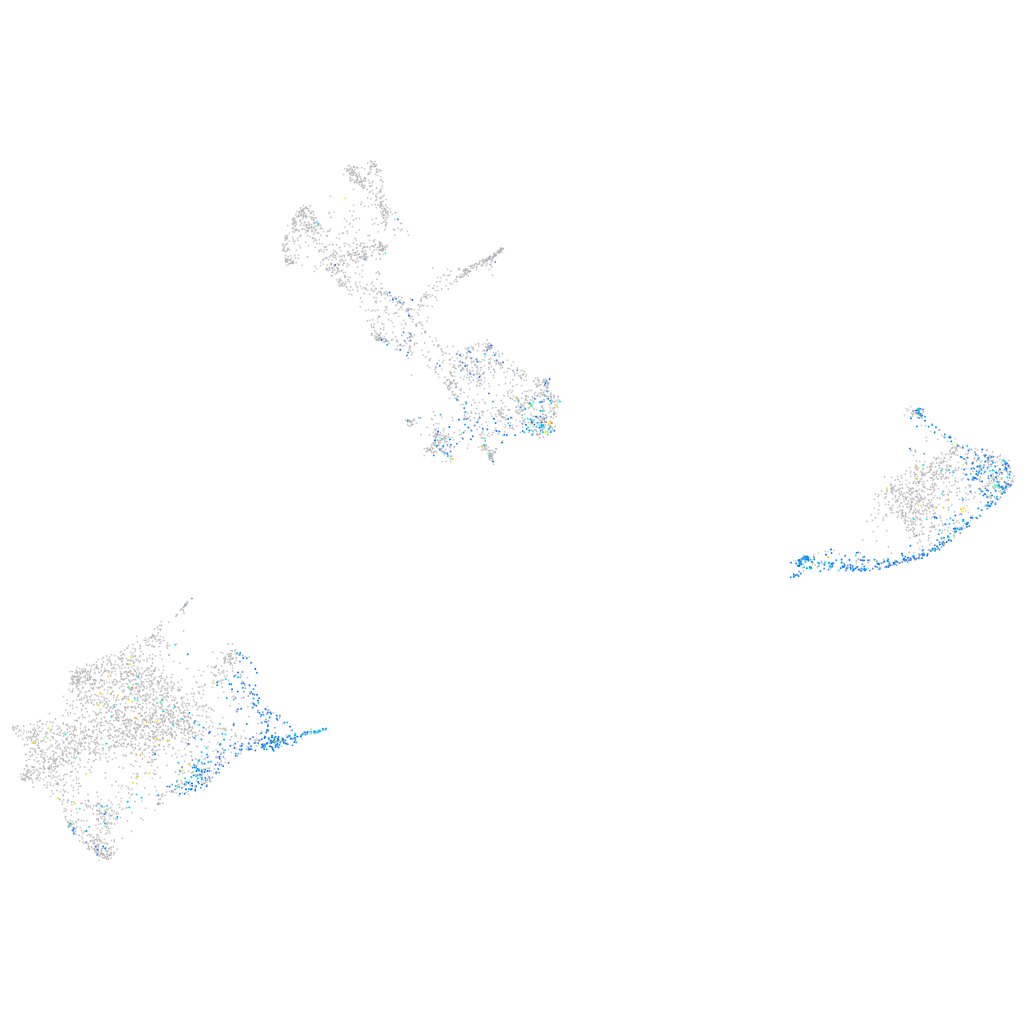
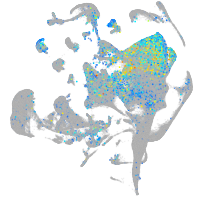
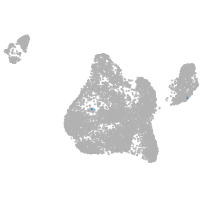
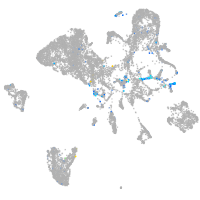
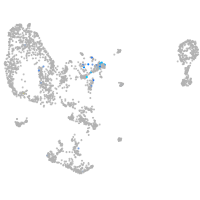
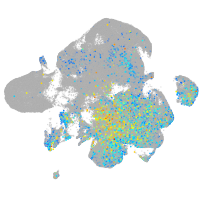
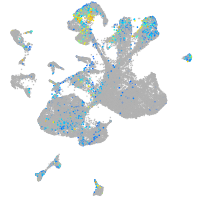
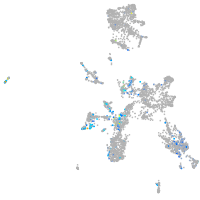
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| SPAG9 | 0.276 | defbl1 | -0.149 |
| mitfa | 0.275 | apoda.1 | -0.148 |
| phlda1 | 0.248 | gpnmb | -0.143 |
| tyr | 0.244 | uraha | -0.143 |
| crestin | 0.243 | si:ch211-256m1.8 | -0.141 |
| pim1 | 0.242 | phyhd1 | -0.138 |
| tfap2e | 0.241 | lypc | -0.137 |
| pcdh10a | 0.234 | mdh1aa | -0.134 |
| tfap2a | 0.234 | pvalb2 | -0.133 |
| lamp1a | 0.231 | si:dkey-197i20.6 | -0.132 |
| ednrab | 0.230 | unm-sa821 | -0.129 |
| msx1b | 0.228 | krt18b | -0.127 |
| zeb2a | 0.227 | pvalb1 | -0.125 |
| XLOC-001964 | 0.226 | si:ch73-89b15.3 | -0.120 |
| itga4 | 0.222 | akap12a | -0.120 |
| canx | 0.220 | alx4b | -0.116 |
| CR383676.1 | 0.220 | ponzr1 | -0.115 |
| xbp1 | 0.220 | alx4a | -0.115 |
| eva1ba | 0.219 | s100v2 | -0.115 |
| tmem243b | 0.214 | sytl2b | -0.113 |
| bace2 | 0.214 | prx | -0.111 |
| opn5 | 0.214 | actc1b | -0.109 |
| atp6ap2 | 0.213 | alx1 | -0.103 |
| glb1l | 0.210 | tpd52l1 | -0.100 |
| atp6ap1b | 0.209 | myof | -0.100 |
| si:zfos-943e10.1 | 0.209 | guk1a | -0.098 |
| sypl2b | 0.209 | cx43 | -0.098 |
| kita | 0.209 | si:dkey-251i10.2 | -0.098 |
| trpm1b | 0.206 | CT737162.3 | -0.097 |
| naga | 0.206 | slc25a36a | -0.096 |
| slc7a5 | 0.205 | fhl2a | -0.096 |
| lman2 | 0.204 | fhl3a | -0.096 |
| pah | 0.204 | tmem179b | -0.095 |
| klf6a | 0.202 | si:ch211-105c13.3 | -0.095 |
| nr2f5 | 0.202 | mylpfa | -0.094 |