protocadherin 9
ZFIN 
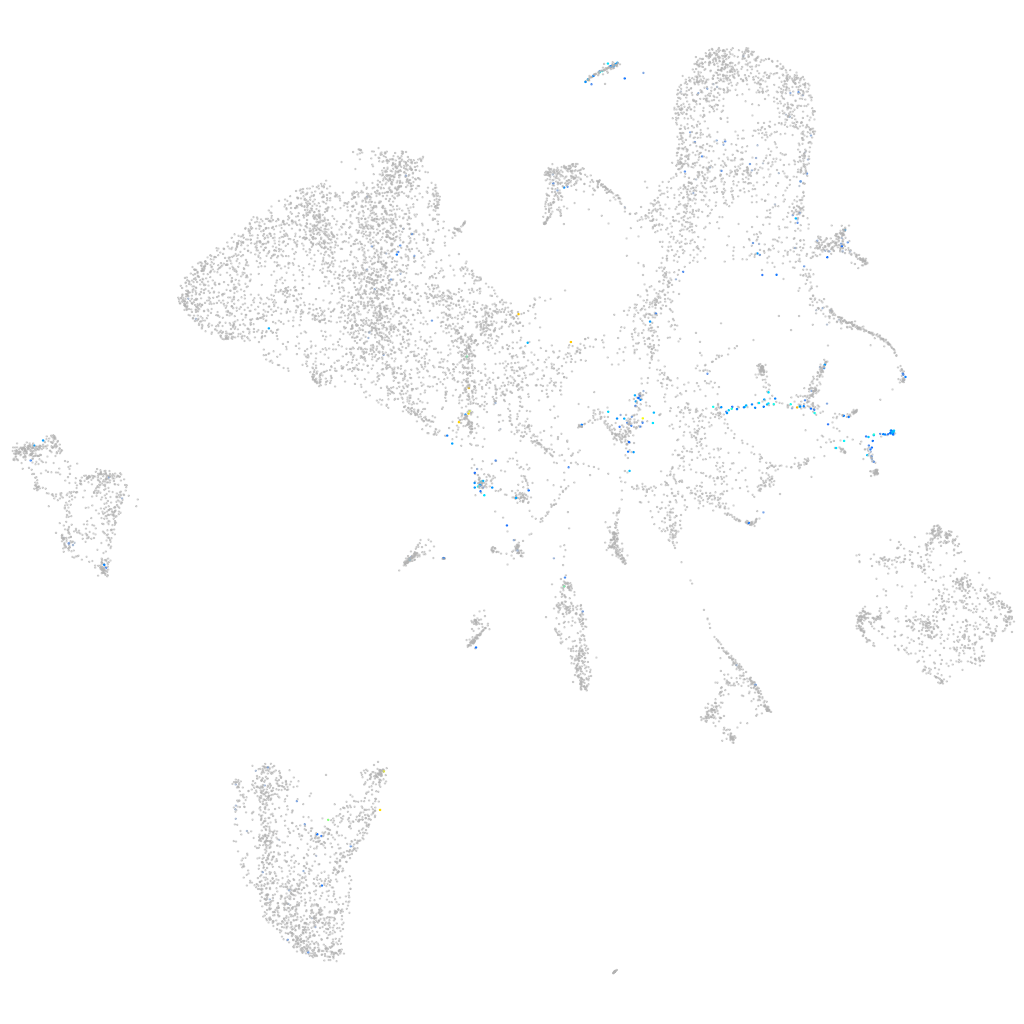
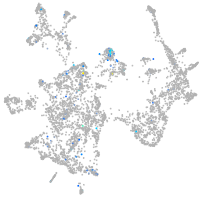
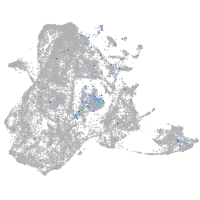
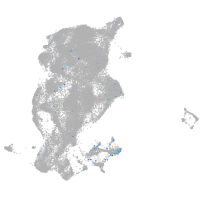
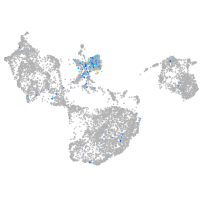
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| sst2 | 0.288 | aldob | -0.111 |
| zgc:195023 | 0.267 | gamt | -0.100 |
| neurod1 | 0.245 | hdlbpa | -0.091 |
| myt1b | 0.233 | gatm | -0.089 |
| scg3 | 0.233 | fbp1b | -0.087 |
| dlb | 0.232 | nupr1b | -0.087 |
| vamp2 | 0.224 | aldh7a1 | -0.087 |
| sst1.2 | 0.220 | gapdh | -0.087 |
| mir7a-1 | 0.217 | mat1a | -0.086 |
| nmba | 0.215 | gstr | -0.082 |
| neurog3 | 0.215 | aldh6a1 | -0.082 |
| insm1b | 0.215 | aqp12 | -0.081 |
| insm1a | 0.214 | scp2a | -0.080 |
| zgc:101731 | 0.203 | cx32.3 | -0.080 |
| mir375-2 | 0.202 | bhmt | -0.077 |
| nkx2.2a | 0.199 | agxtb | -0.076 |
| pax4 | 0.192 | aldh1l1 | -0.076 |
| gpc1a | 0.189 | si:dkey-16p21.8 | -0.076 |
| dll4 | 0.188 | ahcy | -0.075 |
| rnasekb | 0.188 | apoa1b | -0.075 |
| pbx1a | 0.187 | ckba | -0.075 |
| id4 | 0.185 | afp4 | -0.075 |
| cpe | 0.183 | rgrb | -0.075 |
| pdyn | 0.182 | shmt1 | -0.075 |
| sox4b | 0.182 | adka | -0.074 |
| kctd12.2 | 0.181 | agxta | -0.074 |
| mdkb | 0.180 | apoc1 | -0.074 |
| kcnk3b | 0.179 | cpn1 | -0.073 |
| stmn1b | 0.178 | apoc2 | -0.073 |
| tmsb | 0.178 | gpx4a | -0.073 |
| nrcama | 0.178 | mthfd1b | -0.072 |
| gldn | 0.178 | igfbp2a | -0.072 |
| pax6b | 0.177 | sdr16c5b | -0.072 |
| syt1a | 0.174 | suclg1 | -0.072 |
| CABZ01072885.1 | 0.174 | gnmt | -0.071 |