olfactomedin 2a
ZFIN 
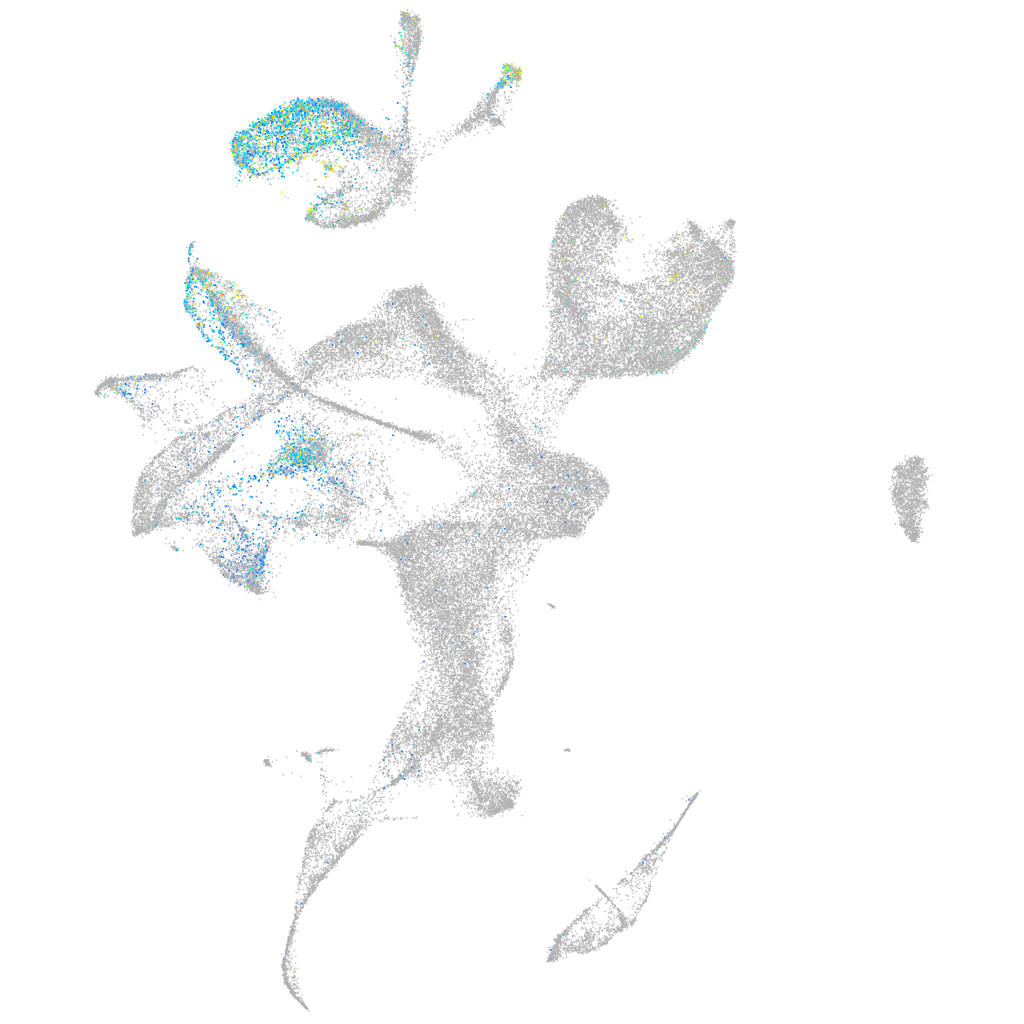
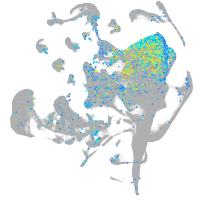
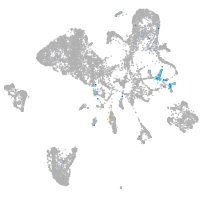
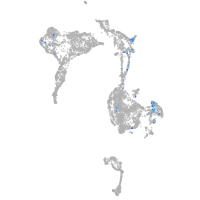
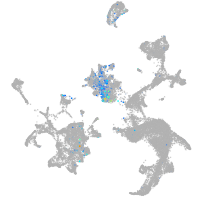
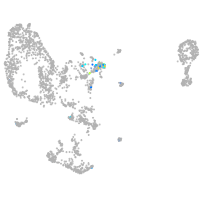
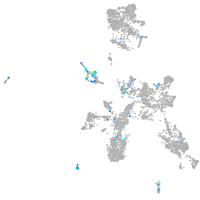
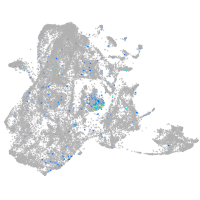
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| stxbp1a | 0.360 | hmgb2a | -0.253 |
| syt1a | 0.353 | pcna | -0.134 |
| ywhag2 | 0.346 | ahcy | -0.134 |
| pax10 | 0.342 | mdka | -0.131 |
| meis2b | 0.332 | stmn1a | -0.130 |
| stx1b | 0.322 | hmga1a | -0.126 |
| rtn1b | 0.318 | rrm1 | -0.125 |
| slc32a1 | 0.311 | msna | -0.121 |
| sv2a | 0.310 | msi1 | -0.121 |
| slc6a1a | 0.309 | fabp7a | -0.121 |
| sncb | 0.309 | lbr | -0.121 |
| gng3 | 0.306 | dut | -0.121 |
| eno2 | 0.305 | mki67 | -0.120 |
| stmn1b | 0.304 | ccnd1 | -0.120 |
| zgc:65894 | 0.301 | nutf2l | -0.120 |
| LOC101886114 | 0.299 | chaf1a | -0.119 |
| slc35g2b | 0.294 | hmgb2b | -0.116 |
| elavl3 | 0.287 | ccna2 | -0.112 |
| celf5a | 0.285 | mcm7 | -0.112 |
| meis2a | 0.284 | tuba8l | -0.111 |
| zgc:153426 | 0.281 | dek | -0.109 |
| elavl4 | 0.281 | banf1 | -0.109 |
| tfap2a | 0.276 | her15.1 | -0.108 |
| sprn | 0.272 | rrm2 | -0.105 |
| ppp1r14ba | 0.272 | selenoh | -0.104 |
| mir181b-3 | 0.271 | cks1b | -0.104 |
| snap25a | 0.270 | lig1 | -0.104 |
| si:ch73-119p20.1 | 0.264 | zgc:110425 | -0.104 |
| nrxn1a | 0.263 | neurod4 | -0.103 |
| syn2a | 0.259 | tuba8l4 | -0.102 |
| tfap2e | 0.259 | mibp | -0.102 |
| rbfox1 | 0.259 | tspan7 | -0.101 |
| nrxn2a | 0.258 | rpa3 | -0.101 |
| gabrb4 | 0.256 | eef1da | -0.101 |
| myt1la | 0.254 | fen1 | -0.100 |