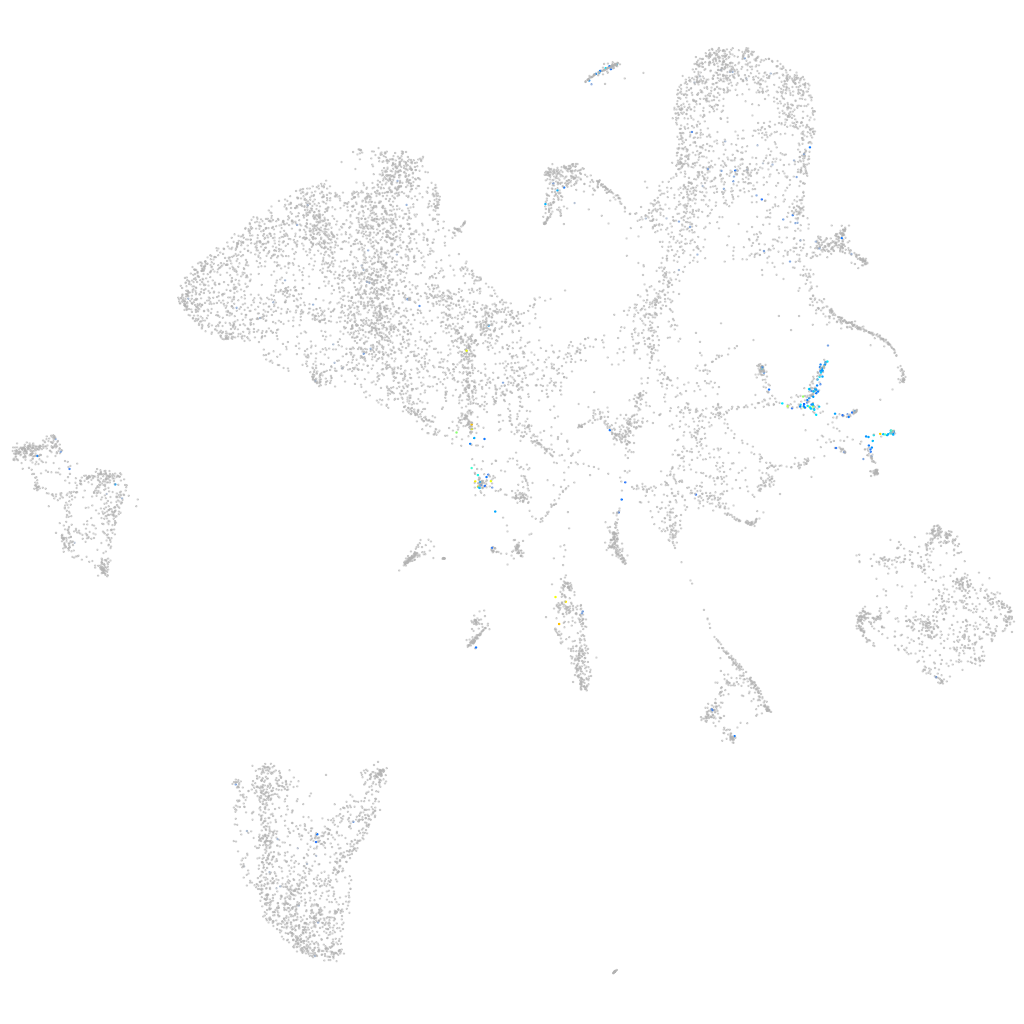
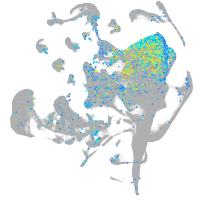
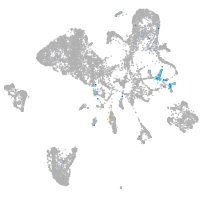
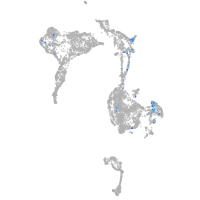
olfactomedin 2a
ZFIN 
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| sst2 | 0.400 | ahcy | -0.096 |
| pdyn | 0.394 | hdlbpa | -0.092 |
| scgn | 0.366 | gamt | -0.092 |
| zgc:195023 | 0.354 | rps20 | -0.091 |
| ghrl | 0.350 | gstr | -0.090 |
| arxa | 0.343 | aldob | -0.090 |
| scg3 | 0.326 | gapdh | -0.089 |
| kctd12.2 | 0.304 | rps12 | -0.088 |
| isl1 | 0.303 | fabp3 | -0.087 |
| CU019662.1 | 0.292 | aldh7a1 | -0.085 |
| syt1a | 0.279 | rpl3 | -0.083 |
| sst1.2 | 0.277 | gatm | -0.082 |
| scg2b | 0.268 | fbp1b | -0.082 |
| nrcama | 0.268 | rpl6 | -0.080 |
| gpc1a | 0.267 | gstt1a | -0.079 |
| c2cd4a | 0.266 | ppa1b | -0.079 |
| adgb | 0.265 | aldh6a1 | -0.079 |
| nmba | 0.264 | apoa4b.1 | -0.079 |
| neurod1 | 0.263 | rpl4 | -0.078 |
| atpv0e2 | 0.263 | eef1da | -0.078 |
| mir7a-1 | 0.260 | scp2a | -0.078 |
| pcsk1nl | 0.258 | cx32.3 | -0.077 |
| rnasekb | 0.257 | glud1b | -0.076 |
| ptger3 | 0.257 | shmt1 | -0.076 |
| vat1 | 0.256 | mgst1.2 | -0.076 |
| pcsk2 | 0.254 | rps6 | -0.076 |
| si:dkey-153k10.9 | 0.250 | ndufa4l | -0.076 |
| atp6v0cb | 0.249 | aqp12 | -0.075 |
| pax6b | 0.249 | mat1a | -0.074 |
| kcnj11 | 0.248 | si:dkey-16p21.8 | -0.073 |
| gldn | 0.241 | gnmt | -0.073 |
| vamp2 | 0.240 | hspd1 | -0.073 |
| tspan7b | 0.240 | dap | -0.073 |
| cplx2 | 0.238 | gstp1 | -0.072 |
| nalcn | 0.238 | agxtb | -0.072 |