N-terminal asparagine amidase
ZFIN 
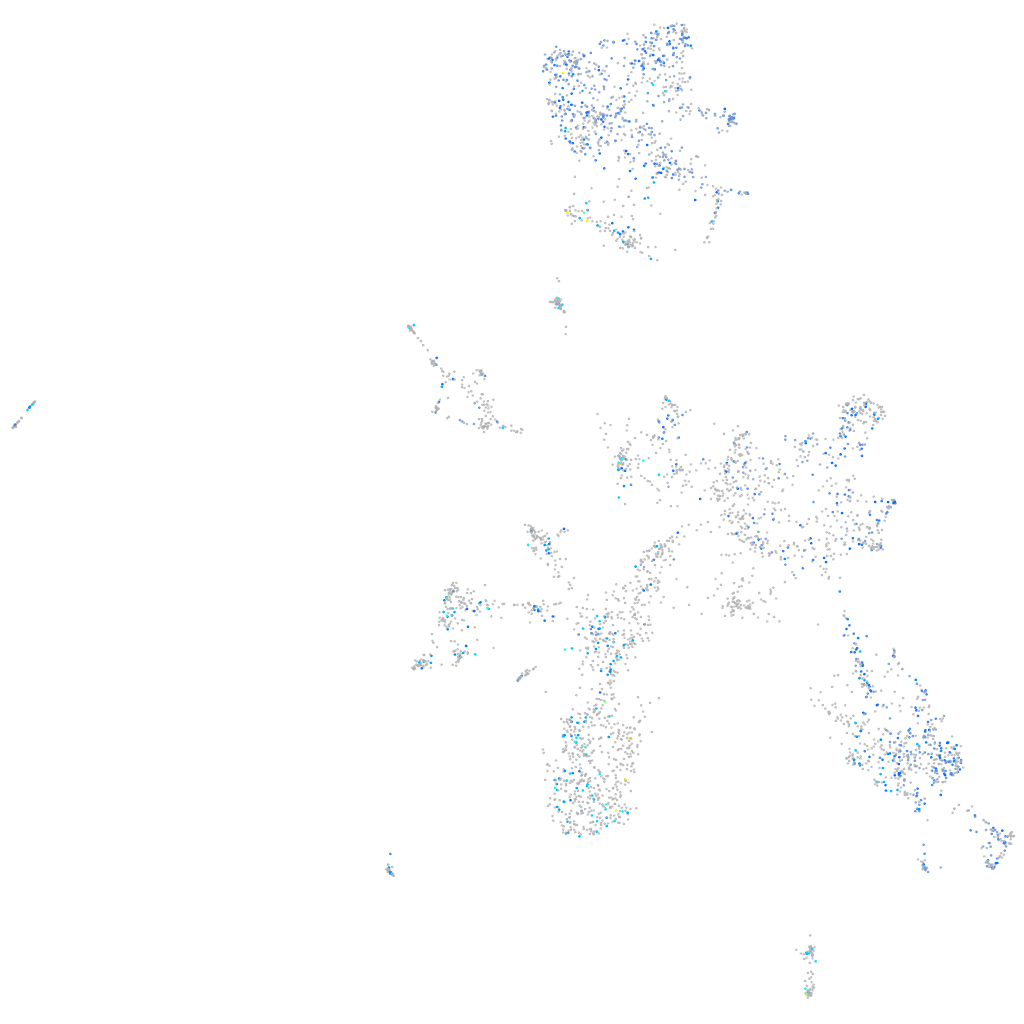
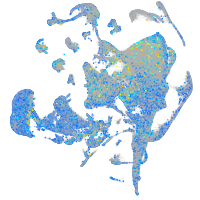
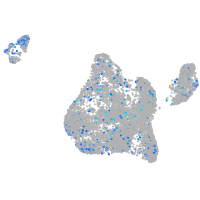
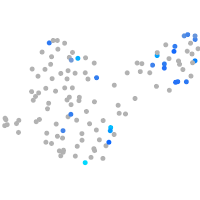
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| capns1b | 0.144 | her4.2 | -0.092 |
| tprg1 | 0.140 | neurod4 | -0.089 |
| ap1s3b | 0.132 | ebf2 | -0.088 |
| flot1b | 0.132 | sox11b | -0.085 |
| GCA | 0.129 | dla | -0.084 |
| degs2 | 0.129 | her15.1 | -0.079 |
| gmds | 0.128 | rps29 | -0.078 |
| cap1 | 0.128 | si:ch211-193l2.6 | -0.077 |
| ftr83 | 0.127 | neurod1 | -0.076 |
| bik | 0.125 | dlx4b | -0.075 |
| tmed2 | 0.125 | zgc:165555.3 | -0.075 |
| oclnb | 0.125 | CU467822.1 | -0.075 |
| tegt | 0.125 | cxcr4b | -0.074 |
| cldne | 0.124 | tp53inp2 | -0.073 |
| si:ch1073-443f11.2 | 0.124 | si:dkey-56m19.5 | -0.072 |
| si:ch211-195b11.3 | 0.124 | srrm4 | -0.071 |
| chmp5b | 0.124 | XLOC-003692 | -0.068 |
| cd9b | 0.123 | stmn1a | -0.068 |
| anxa11b | 0.122 | hist1h4l | -0.068 |
| zgc:92313 | 0.122 | sox11a | -0.068 |
| gbgt1l3 | 0.121 | rpl38 | -0.067 |
| zgc:158343 | 0.121 | rrm2 | -0.067 |
| rassf7b | 0.120 | ccna2 | -0.067 |
| elovl1b | 0.120 | inka1a | -0.067 |
| vamp8 | 0.120 | elavl3 | -0.066 |
| cx35.4 | 0.120 | her4.1 | -0.066 |
| dhrs13l1 | 0.120 | ptmab | -0.065 |
| litaf | 0.120 | zgc:110216 | -0.063 |
| mvp | 0.118 | emx3 | -0.063 |
| mpzl2b | 0.118 | rbbp4 | -0.063 |
| krt4 | 0.118 | lfng | -0.063 |
| spint1a | 0.118 | mibp | -0.061 |
| zgc:101744 | 0.117 | spc25 | -0.061 |
| si:ch73-347e22.8 | 0.117 | abhd6a | -0.060 |
| calm1b | 0.117 | hist1h2a11 | -0.060 |