"NTPase, KAP family P-loop domain containing 1"
ZFIN 
Other cell groups


all cells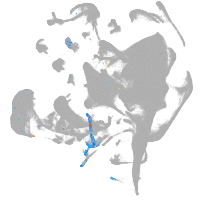 |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros 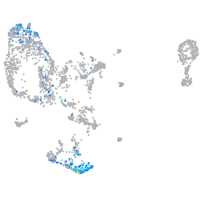 |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| trim35-12 | 0.464 | si:ch73-1a9.3 | -0.172 |
| slc7a8a | 0.459 | marcksl1b | -0.169 |
| atp1a1a.3 | 0.457 | hspb1 | -0.164 |
| si:dkey-36i7.3 | 0.456 | nucks1a | -0.155 |
| etf1a | 0.448 | hnrnpabb | -0.154 |
| slc9a3.2 | 0.447 | marcksb | -0.151 |
| soul2 | 0.430 | eif5a2 | -0.148 |
| ca4c | 0.424 | wu:fb97g03 | -0.144 |
| zgc:158423 | 0.416 | si:ch73-281n10.2 | -0.140 |
| glsb | 0.410 | nono | -0.139 |
| aspg | 0.408 | tubb2b | -0.138 |
| rhcgb | 0.397 | apoeb | -0.138 |
| atp1a1a.5 | 0.396 | msna | -0.133 |
| rps6ka5 | 0.386 | ilf3b | -0.131 |
| mal2 | 0.379 | ptmab | -0.130 |
| ppp1r1b | 0.378 | hmgb1b | -0.129 |
| slc12a10.3 | 0.377 | hmgb2b | -0.129 |
| glud1a | 0.373 | hnrnpaba | -0.128 |
| fabp2 | 0.373 | cx43.4 | -0.128 |
| me3 | 0.371 | hnrnph1l | -0.128 |
| tmem176l.4 | 0.364 | syncrip | -0.127 |
| slc3a2a | 0.363 | akap12b | -0.126 |
| si:ch211-153b23.5 | 0.358 | acin1a | -0.126 |
| alas1 | 0.355 | hmga1a | -0.125 |
| emx1 | 0.352 | fbl | -0.123 |
| rnf183 | 0.350 | apoc1 | -0.122 |
| plcd3a | 0.348 | lmo2 | -0.122 |
| phlda2 | 0.347 | anp32e | -0.121 |
| rhbg | 0.346 | zgc:110425 | -0.121 |
| si:ch211-79k12.1 | 0.345 | seta | -0.121 |
| BX000438.2 | 0.343 | setb | -0.120 |
| agr2 | 0.343 | h3f3d | -0.119 |
| bsnd | 0.339 | id3 | -0.119 |
| tspan13a | 0.338 | hnrnpa1a | -0.119 |
| gdpd3a | 0.338 | si:rp71-17i16.6 | -0.117 |




