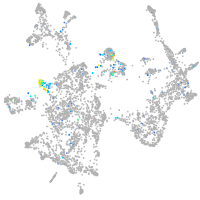
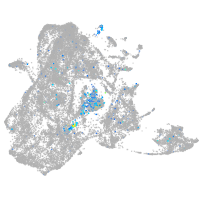
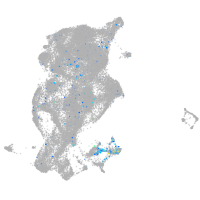
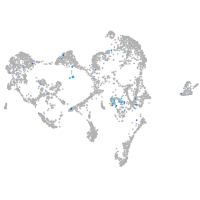
neuronal differentiation 1
ZFIN 
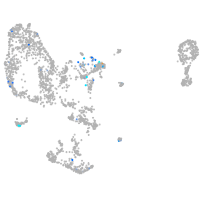
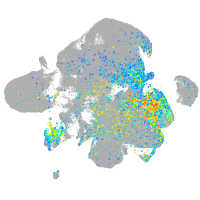
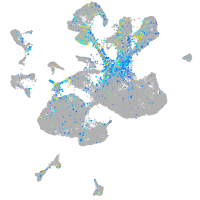
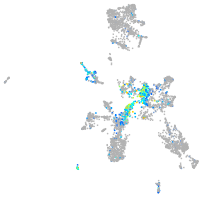
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| neurod2 | 0.309 | gpr143 | -0.117 |
| zbtb18 | 0.257 | tspan36 | -0.113 |
| gpm6aa | 0.249 | smim29 | -0.111 |
| otx5 | 0.246 | syngr1a | -0.109 |
| snap25b | 0.242 | cst14a.2 | -0.101 |
| rnasekb | 0.242 | gstp1 | -0.096 |
| nova2 | 0.240 | rabl6b | -0.096 |
| syt5b | 0.239 | pttg1ipb | -0.095 |
| ptmaa | 0.237 | rnaseka | -0.094 |
| neurod6b | 0.231 | pcbd1 | -0.091 |
| elavl3 | 0.229 | paics | -0.090 |
| scrt2 | 0.228 | impdh1b | -0.090 |
| stmn1b | 0.227 | qdpra | -0.090 |
| epb41a | 0.225 | rab32a | -0.090 |
| crx | 0.224 | C12orf75 | -0.089 |
| CU467822.1 | 0.223 | gart | -0.089 |
| tuba1c | 0.219 | akr1b1 | -0.088 |
| sypb | 0.219 | slc2a15a | -0.087 |
| nfixb | 0.213 | si:dkey-21a6.5 | -0.087 |
| LOC103909284 | 0.212 | zgc:110239 | -0.087 |
| foxg1b | 0.212 | actb2 | -0.086 |
| arl3l1 | 0.211 | eno3 | -0.086 |
| atp1a3a | 0.210 | atox1 | -0.085 |
| cspg5b | 0.210 | slc45a2 | -0.084 |
| rs1a | 0.208 | rab34b | -0.084 |
| cadm3 | 0.208 | ctsba | -0.083 |
| ptmab | 0.208 | adi1 | -0.083 |
| si:ch73-256g18.2 | 0.205 | agtrap | -0.082 |
| atp6v0cb | 0.204 | rab38 | -0.082 |
| insm1a | 0.204 | zgc:165573 | -0.082 |
| elovl4b | 0.204 | prdx1 | -0.081 |
| hmgb1b | 0.204 | zgc:110591 | -0.081 |
| nptnb | 0.204 | LOC103910009 | -0.080 |
| si:ch211-222l21.1 | 0.200 | gmps | -0.080 |
| gngt2a | 0.200 | ahnak | -0.080 |