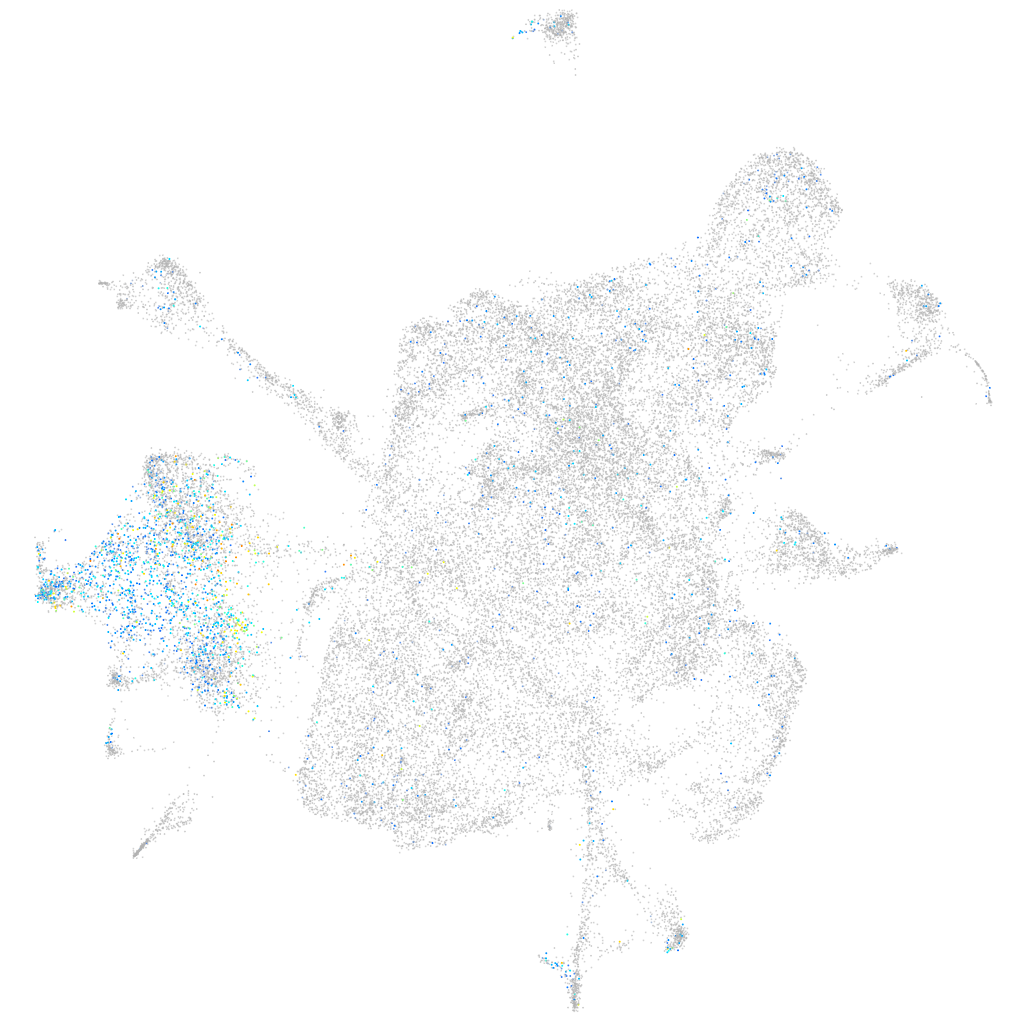
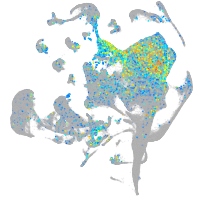
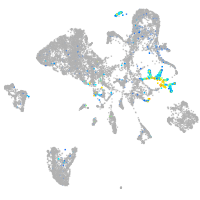
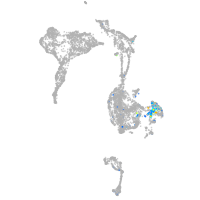
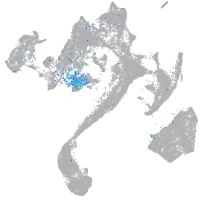
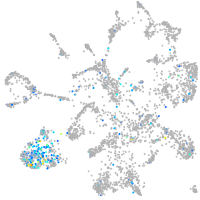
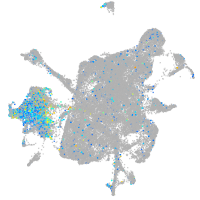
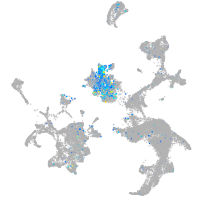
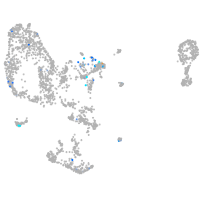
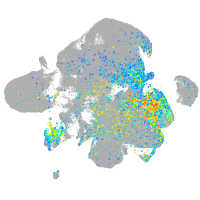
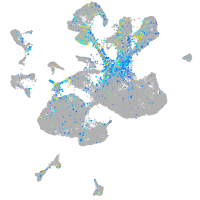
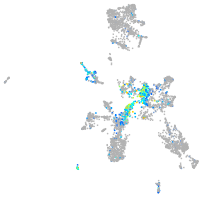
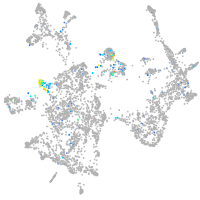
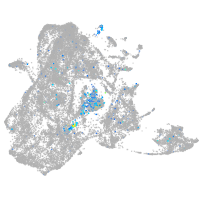
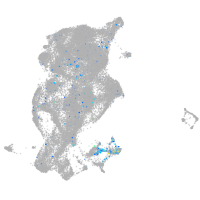
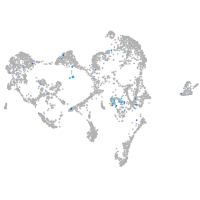
neuronal differentiation 1
ZFIN 
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| neurod2 | 0.332 | pmp22a | -0.104 |
| elavl3 | 0.294 | tpm4a | -0.087 |
| gpm6aa | 0.283 | krt18a.1 | -0.078 |
| stmn1b | 0.271 | mfap2 | -0.077 |
| tuba1c | 0.261 | rpl9 | -0.071 |
| zbtb18 | 0.259 | rps8a | -0.070 |
| myt1a | 0.250 | sdc4 | -0.070 |
| insm1a | 0.250 | krt8 | -0.069 |
| rtn1a | 0.247 | eef1a1l1 | -0.067 |
| nova2 | 0.242 | rps2 | -0.067 |
| ckbb | 0.240 | rpl15 | -0.066 |
| neurod6b | 0.237 | rpsa | -0.065 |
| bhlhe22 | 0.231 | rpl12 | -0.065 |
| sypb | 0.231 | twist1a | -0.064 |
| scrt2 | 0.230 | rps27a | -0.063 |
| epb41a | 0.228 | fkbp9 | -0.063 |
| cadm3 | 0.228 | id3 | -0.062 |
| syt5b | 0.227 | her6 | -0.062 |
| insm1b | 0.225 | rpl5a | -0.061 |
| gng3 | 0.223 | rpl18 | -0.061 |
| CU467822.1 | 0.222 | crtap | -0.061 |
| snap25b | 0.221 | rps24 | -0.061 |
| atp6v0cb | 0.218 | rpl18a | -0.060 |
| si:ch211-137a8.4 | 0.218 | lin28a | -0.058 |
| gpm6ab | 0.213 | rps19 | -0.058 |
| tuba1a | 0.212 | tpt1 | -0.057 |
| elavl4 | 0.212 | rpl26 | -0.057 |
| neurod4 | 0.211 | rpl24 | -0.057 |
| rtn1b | 0.210 | colec12 | -0.057 |
| tmsb | 0.210 | rpl3 | -0.056 |
| otx5 | 0.208 | rps6 | -0.056 |
| rs1a | 0.206 | fkbp14 | -0.055 |
| golga7ba | 0.203 | fkbp7 | -0.055 |
| bhlhe23 | 0.201 | rps14 | -0.055 |
| rnasekb | 0.200 | rps9 | -0.054 |