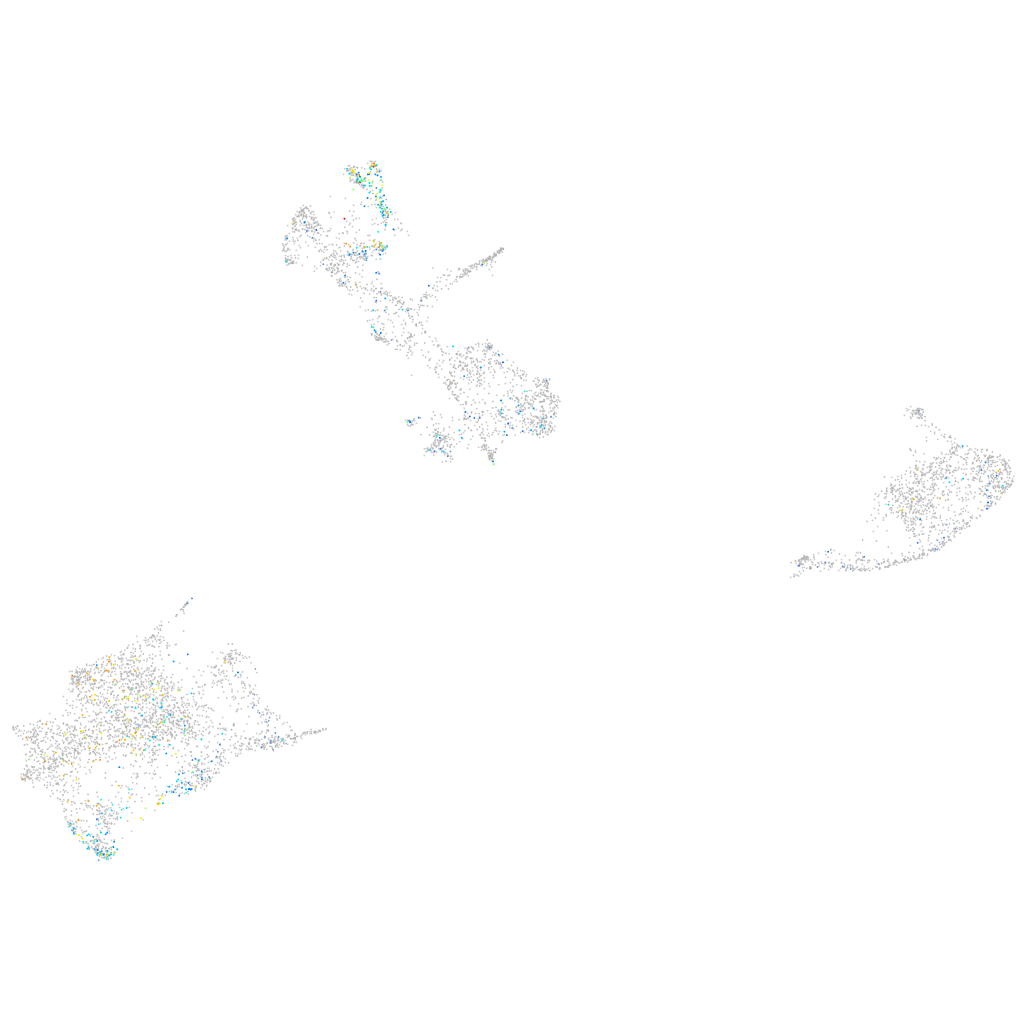
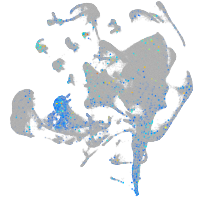
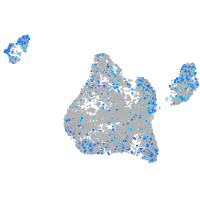
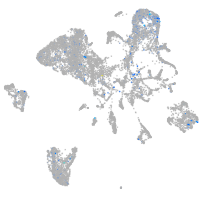
myosin VAb
ZFIN 
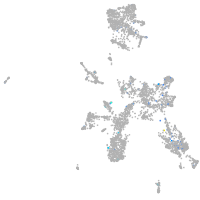
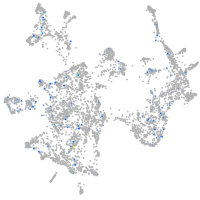
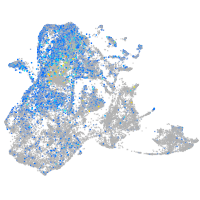
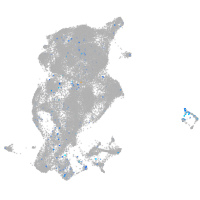
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| CU855878.1 | 0.197 | pmela | -0.090 |
| FO704779.1 | 0.197 | tyrp1b | -0.088 |
| id2b | 0.184 | zgc:91968 | -0.087 |
| dpep2 | 0.176 | ptmab | -0.086 |
| kitlga | 0.171 | tyrp1a | -0.083 |
| itga6b | 0.171 | hmga1a | -0.082 |
| rgs4 | 0.171 | si:ch211-222l21.1 | -0.082 |
| sort1a | 0.168 | dct | -0.080 |
| lgals2a | 0.149 | tubb2b | -0.079 |
| mdh1aa | 0.145 | si:ch73-389b16.1 | -0.071 |
| sesn1 | 0.142 | oca2 | -0.071 |
| tcirg1a | 0.142 | tyr | -0.070 |
| si:dkey-44g17.6 | 0.142 | marcksl1a | -0.070 |
| clec3ba | 0.141 | hmgb2b | -0.070 |
| vip | 0.140 | slc24a5 | -0.068 |
| gpnmb | 0.137 | khdrbs1a | -0.067 |
| zgc:110789 | 0.136 | tuba1a | -0.067 |
| alx4b | 0.132 | h3f3a | -0.066 |
| phyhd1 | 0.132 | mtbl | -0.065 |
| cln3 | 0.130 | tfap2a | -0.064 |
| nucb2b | 0.130 | kita | -0.064 |
| gpx2 | 0.129 | pcna | -0.063 |
| pde9al | 0.129 | marcksb | -0.063 |
| sytl2a | 0.128 | slc39a10 | -0.063 |
| sod1 | 0.128 | hnrnpaba | -0.063 |
| rnd3b | 0.126 | prkar1b | -0.062 |
| slc2a15a | 0.125 | gstt1a | -0.062 |
| cox4i2 | 0.124 | si:dkey-151g10.6 | -0.061 |
| si:ch211-38m6.6 | 0.124 | snrpb | -0.059 |
| pfas | 0.124 | apex1 | -0.058 |
| dpysl3 | 0.124 | fen1 | -0.058 |
| lypc | 0.124 | hnrnpa0b | -0.058 |
| akap12a | 0.123 | snrpe | -0.058 |
| si:ch211-133l5.7 | 0.123 | setb | -0.058 |
| glulb | 0.123 | ptges3b | -0.056 |