macrophage stimulating 1 receptor a
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle 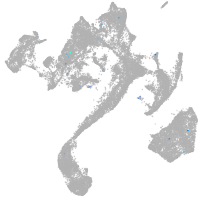 |
mural cells / non-skeletal muscle  |
mesenchyme 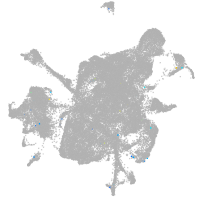 |
hematopoietic / vasculature  |
pronephros 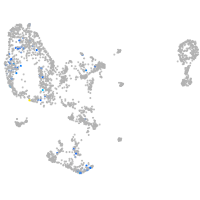 |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis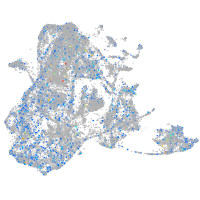 |
periderm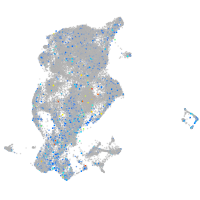 |
fin |
ionocytes / mucous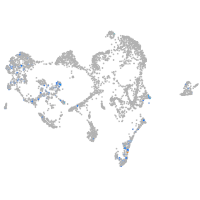 |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| s100a10a | 0.248 | bhmt | -0.168 |
| zgc:158846 | 0.245 | aqp12 | -0.150 |
| krt92 | 0.244 | apoc1 | -0.127 |
| serpinb1 | 0.243 | agxtb | -0.118 |
| anxa2b | 0.240 | ppdpfa | -0.116 |
| serpinb1l3 | 0.238 | fabp3 | -0.113 |
| itpk1a | 0.238 | kng1 | -0.109 |
| lrrc66 | 0.236 | rtn1a | -0.108 |
| wu:fb59d01 | 0.233 | gatm | -0.104 |
| eps8l3b | 0.232 | apoda.2 | -0.104 |
| si:dkey-283b1.6 | 0.228 | apoa2 | -0.103 |
| gsta.1 | 0.226 | zgc:112160 | -0.103 |
| atp1a1a.4 | 0.224 | prss59.1 | -0.102 |
| atp1b1a | 0.224 | si:ch211-240l19.5 | -0.102 |
| bin2a | 0.223 | prss1 | -0.102 |
| eps8l3a | 0.223 | sycn.2 | -0.102 |
| si:dkey-36i7.3 | 0.222 | cel.1 | -0.102 |
| ezra | 0.221 | cpb1 | -0.101 |
| nccrp1 | 0.220 | ela2 | -0.101 |
| GCA | 0.218 | fep15 | -0.101 |
| tagln2 | 0.215 | ela2l | -0.101 |
| plac8.1 | 0.213 | CELA1 (1 of many) | -0.101 |
| gnpda1 | 0.213 | cpa5 | -0.101 |
| myo15b | 0.211 | ttc36 | -0.100 |
| gstp1 | 0.210 | tfa | -0.100 |
| krt4 | 0.210 | cel.2 | -0.100 |
| eppk1 | 0.209 | ctrb1 | -0.100 |
| gstm.3 | 0.208 | prss59.2 | -0.100 |
| degs2 | 0.208 | erp27 | -0.100 |
| zgc:172079 | 0.208 | rbp2b | -0.099 |
| lamb2l | 0.207 | ela3l | -0.099 |
| lxn | 0.206 | serpina1 | -0.099 |
| phlda2 | 0.205 | pdia2 | -0.099 |
| pfn2 | 0.205 | serpina1l | -0.099 |
| si:ch211-137i24.10 | 0.204 | zgc:136461 | -0.099 |




