macrophage stimulating 1 receptor a
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis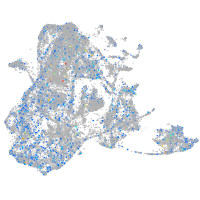 |
periderm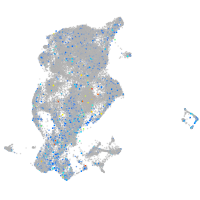 |
fin |
ionocytes / mucous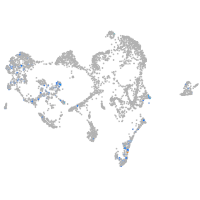 |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| LOC103910418 | 0.092 | rpl38 | -0.062 |
| myh9a | 0.092 | rpl39 | -0.061 |
| epcam | 0.090 | rps29 | -0.060 |
| pdlim1 | 0.089 | rpl36a | -0.058 |
| tagln2 | 0.086 | rps21 | -0.057 |
| ywhaz | 0.085 | rpl37 | -0.057 |
| sdr16c5a | 0.085 | si:dkey-151g10.6 | -0.056 |
| myl12.1 | 0.084 | rpl31 | -0.051 |
| tpm3 | 0.083 | rplp1 | -0.050 |
| st14a | 0.083 | rpl29 | -0.050 |
| klf6a | 0.082 | rpl36 | -0.047 |
| LOC110437833 | 0.081 | rps28 | -0.045 |
| abracl | 0.081 | rpl34 | -0.043 |
| ahnak | 0.080 | zgc:114188 | -0.042 |
| cdh1 | 0.080 | rplp2l | -0.041 |
| actb1 | 0.079 | rps25 | -0.041 |
| tmem54a | 0.079 | rpl28 | -0.039 |
| cldni | 0.078 | rpl35 | -0.038 |
| tpm4a | 0.078 | si:dkey-183i3.5 | -0.038 |
| jupa | 0.078 | stmn1a | -0.038 |
| lama5 | 0.077 | rps24 | -0.038 |
| zgc:171775 | 0.077 | six1b | -0.038 |
| myo1eb | 0.076 | zgc:136930 | -0.037 |
| cnn2 | 0.076 | tuba1c | -0.037 |
| crtap | 0.076 | rps12 | -0.036 |
| col7a1l | 0.076 | rpl35a | -0.035 |
| iqgap1 | 0.075 | ahcy | -0.035 |
| arpc3 | 0.075 | rpl23a | -0.034 |
| bcam | 0.075 | rpl27 | -0.034 |
| arpc5a | 0.074 | eef1b2 | -0.034 |
| tln1 | 0.074 | rps19 | -0.032 |
| arpc4l | 0.073 | six1a | -0.031 |
| zgc:162730 | 0.073 | eif4ebp3 | -0.031 |
| fkbp14 | 0.073 | atp5f1e | -0.031 |
| actn1 | 0.072 | rps26l | -0.031 |