macrophage stimulating 1 receptor a
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle 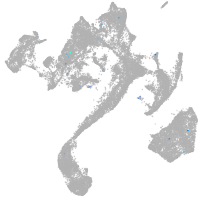 |
mural cells / non-skeletal muscle  |
mesenchyme 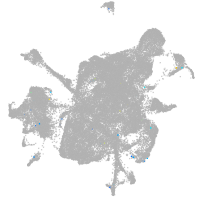 |
hematopoietic / vasculature  |
pronephros 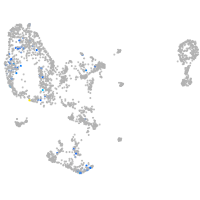 |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis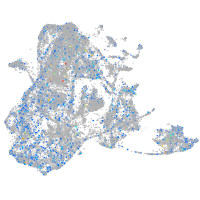 |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| tmsb1 | 0.209 | hmgb1b | -0.094 |
| epcam | 0.199 | tuba1c | -0.086 |
| cfl1l | 0.195 | nova2 | -0.085 |
| myh9a | 0.188 | marcksb | -0.083 |
| arhgef5 | 0.185 | si:dkey-56m19.5 | -0.079 |
| s100a10b | 0.181 | hmgb3a | -0.077 |
| cdh1 | 0.179 | si:ch211-137a8.4 | -0.077 |
| s100v1 | 0.179 | tuba1a | -0.077 |
| cd9b | 0.177 | gpm6aa | -0.075 |
| pfn1 | 0.177 | mdkb | -0.073 |
| lgals3b | 0.176 | si:ch211-288g17.3 | -0.072 |
| spint1a | 0.174 | hnrnpa0a | -0.071 |
| cldni | 0.173 | rtn1a | -0.071 |
| krt4 | 0.172 | ckbb | -0.069 |
| dnase1l4.1 | 0.171 | marcksl1a | -0.069 |
| cebpb | 0.170 | si:dkey-276j7.1 | -0.068 |
| krt92 | 0.170 | elavl3 | -0.067 |
| gsta.1 | 0.166 | fabp3 | -0.066 |
| capns1a | 0.164 | gpm6ab | -0.065 |
| mapk12a | 0.164 | stmn1b | -0.064 |
| gpa33a | 0.162 | zc4h2 | -0.063 |
| cldn1 | 0.161 | si:ch211-133n4.4 | -0.062 |
| tmem54a | 0.160 | tubb5 | -0.062 |
| gsnb | 0.158 | atp6v0cb | -0.060 |
| hbegfa | 0.158 | tmeff1b | -0.060 |
| itgb1b.1 | 0.158 | fscn1a | -0.059 |
| f11r.1 | 0.157 | jpt1b | -0.059 |
| jupa | 0.157 | h2afva | -0.058 |
| anxa1a | 0.156 | chd4a | -0.057 |
| si:ch211-166a6.5 | 0.155 | rnasekb | -0.057 |
| spaca4l | 0.155 | vdac1 | -0.057 |
| tmem238a | 0.155 | cspg5a | -0.056 |
| zgc:153284 | 0.155 | epb41a | -0.056 |
| cldn7b | 0.154 | smarce1 | -0.056 |
| sh3d21 | 0.154 | fam168a | -0.055 |




