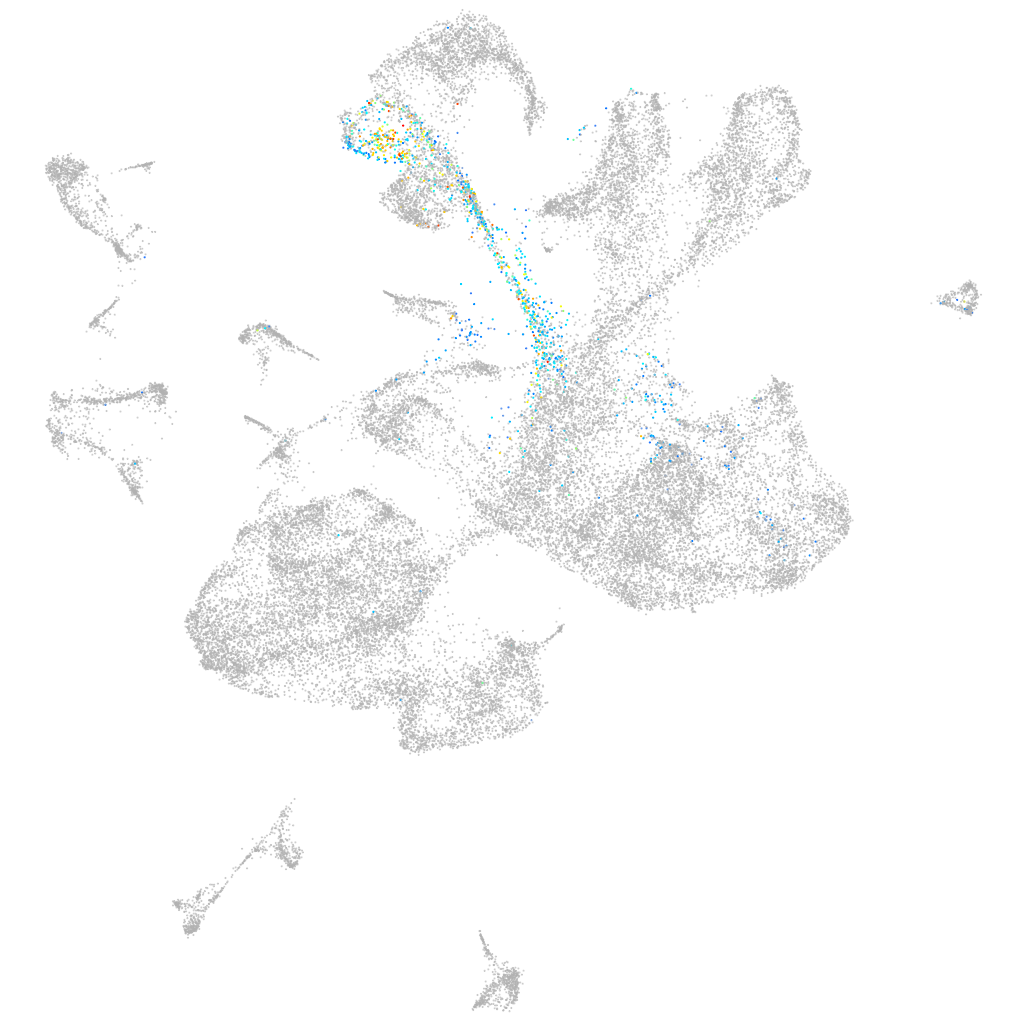
motor neuron and pancreas homeobox 2a
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia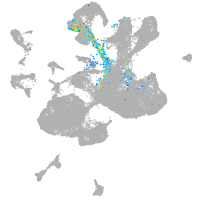 |
eye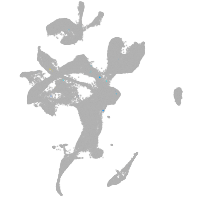 |
taste / olfactory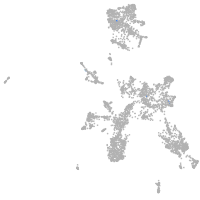 |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells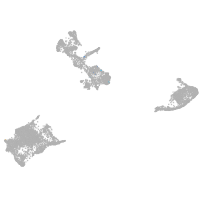 |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| mnx2b | 0.451 | id1 | -0.121 |
| lhx4 | 0.360 | sparc | -0.109 |
| LHX3 | 0.349 | mdkb | -0.088 |
| mnx1 | 0.331 | fabp7a | -0.087 |
| slc18a3a | 0.278 | atp1a1b | -0.084 |
| tuba8l3 | 0.272 | cd82a | -0.079 |
| tac1 | 0.271 | slc1a2b | -0.077 |
| slc5a7a | 0.246 | mdka | -0.077 |
| si:ch211-247j9.1 | 0.236 | XLOC-003690 | -0.075 |
| cxcr4b | 0.234 | tuba8l4 | -0.074 |
| alcama | 0.231 | fosab | -0.074 |
| LOC100537369 | 0.230 | qki2 | -0.074 |
| chga | 0.218 | sox2 | -0.074 |
| isl1 | 0.211 | boc | -0.074 |
| isl2a | 0.209 | atp1b1a | -0.073 |
| gfra3 | 0.205 | cldn5a | -0.072 |
| si:ch211-263k4.2 | 0.204 | zfp36l1a | -0.072 |
| myt1b | 0.203 | pax3a | -0.072 |
| cntn2 | 0.198 | msi1 | -0.071 |
| sv2c | 0.197 | pcna | -0.071 |
| chata | 0.197 | tuba8l | -0.070 |
| ret | 0.194 | her9 | -0.070 |
| l1cama | 0.187 | selenoh | -0.070 |
| ache | 0.185 | her2 | -0.069 |
| slit3 | 0.185 | msna | -0.069 |
| aclya | 0.185 | atp1b4 | -0.068 |
| ptprfa | 0.184 | id3 | -0.068 |
| elavl3 | 0.178 | cspg5a | -0.067 |
| CABZ01074363.1 | 0.177 | sdc4 | -0.067 |
| rab3da | 0.174 | psat1 | -0.066 |
| ribc1 | 0.173 | celf2 | -0.066 |
| flrt1b | 0.172 | COX7A2 (1 of many) | -0.066 |
| agrn | 0.171 | tpm4a | -0.066 |
| SLC35D3 | 0.169 | sox3 | -0.066 |
| chodl | 0.169 | ccnd1 | -0.065 |