low density lipoprotein receptor adaptor protein 1a
ZFIN 
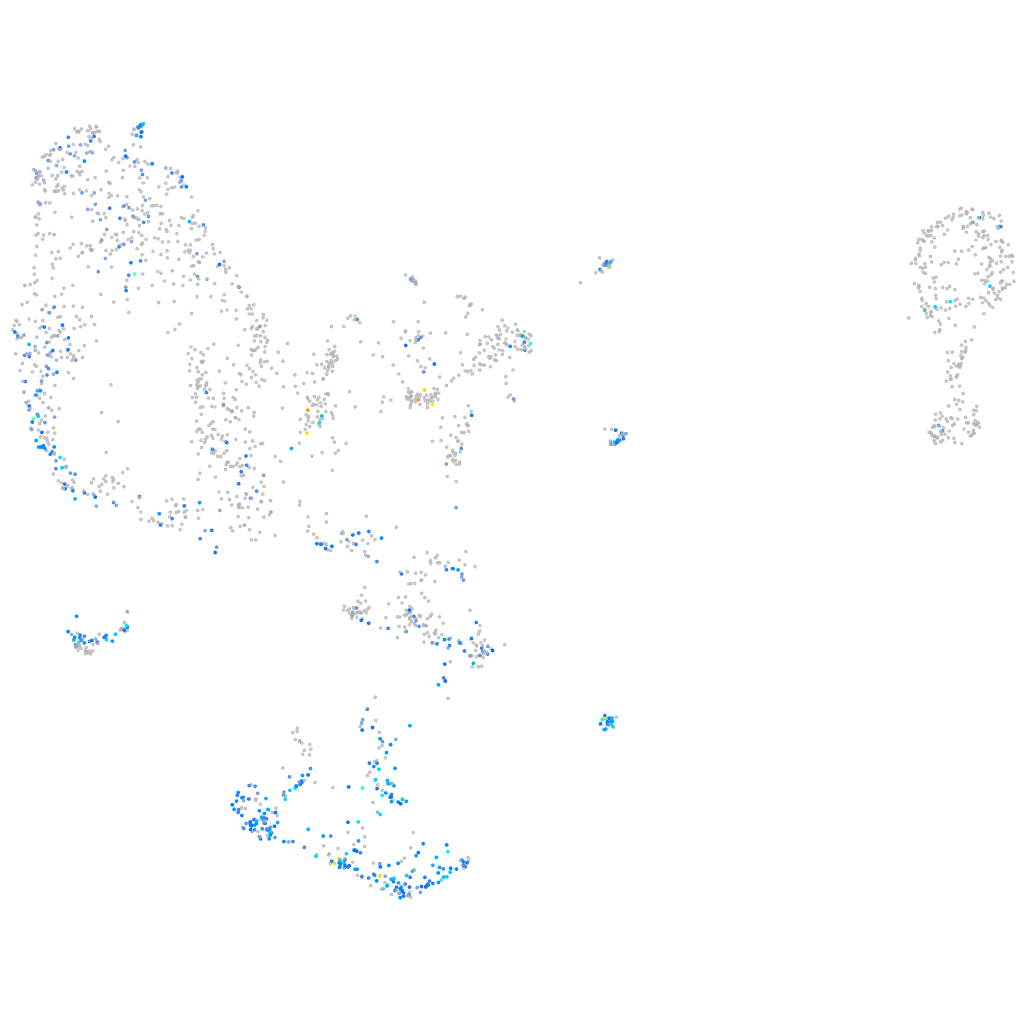
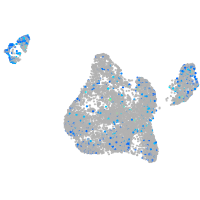
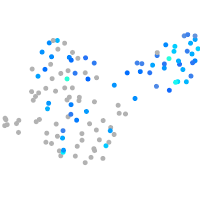
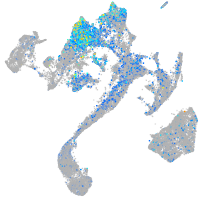
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| phlda2 | 0.461 | aqp8b | -0.237 |
| clcnk | 0.430 | dab2 | -0.225 |
| bsnd | 0.424 | zgc:136493 | -0.211 |
| malb | 0.414 | si:dkey-28n18.9 | -0.201 |
| rnf183 | 0.405 | fxyd6l | -0.198 |
| cd63 | 0.402 | lrp2a | -0.196 |
| si:ch73-359m17.9 | 0.399 | acy3.2 | -0.187 |
| zgc:158423 | 0.397 | slc4a4a | -0.185 |
| mal2 | 0.384 | add3a | -0.182 |
| ccl19a.1 | 0.384 | si:dkey-103j14.5 | -0.170 |
| cldnh | 0.374 | slc6a19a.1 | -0.166 |
| tmem72 | 0.374 | grhprb | -0.166 |
| hoxb8a | 0.365 | nit2 | -0.164 |
| gdpd3a | 0.362 | slc26a1 | -0.164 |
| elf3 | 0.360 | si:dkey-194e6.1 | -0.161 |
| sfxn3 | 0.354 | nrip2 | -0.161 |
| cxcl20 | 0.337 | mafa | -0.159 |
| gucy2c | 0.334 | gpt2l | -0.159 |
| mylk5 | 0.333 | npm1a | -0.158 |
| f11r.1 | 0.333 | aldob | -0.158 |
| agr2 | 0.332 | zgc:101040 | -0.157 |
| trim35-12 | 0.329 | pnp6 | -0.157 |
| slc25a48 | 0.328 | elovl5 | -0.157 |
| soul2 | 0.327 | fbp1b | -0.157 |
| atp1a1a.3 | 0.326 | slc20a1a | -0.157 |
| degs2 | 0.325 | wu:fb97g03 | -0.155 |
| tagln2 | 0.322 | cubn | -0.155 |
| ctgfa | 0.321 | si:ch211-132b12.2 | -0.154 |
| alas1 | 0.319 | lrpap1 | -0.150 |
| me3 | 0.314 | fbl | -0.149 |
| gadd45bb | 0.312 | marcksl1b | -0.148 |
| etf1a | 0.311 | epdl2 | -0.147 |
| zgc:162730 | 0.310 | cx43.4 | -0.146 |
| si:dkey-36i7.3 | 0.309 | ptmab | -0.145 |
| quo | 0.309 | marcksb | -0.144 |