low density lipoprotein receptor adaptor protein 1a
ZFIN 
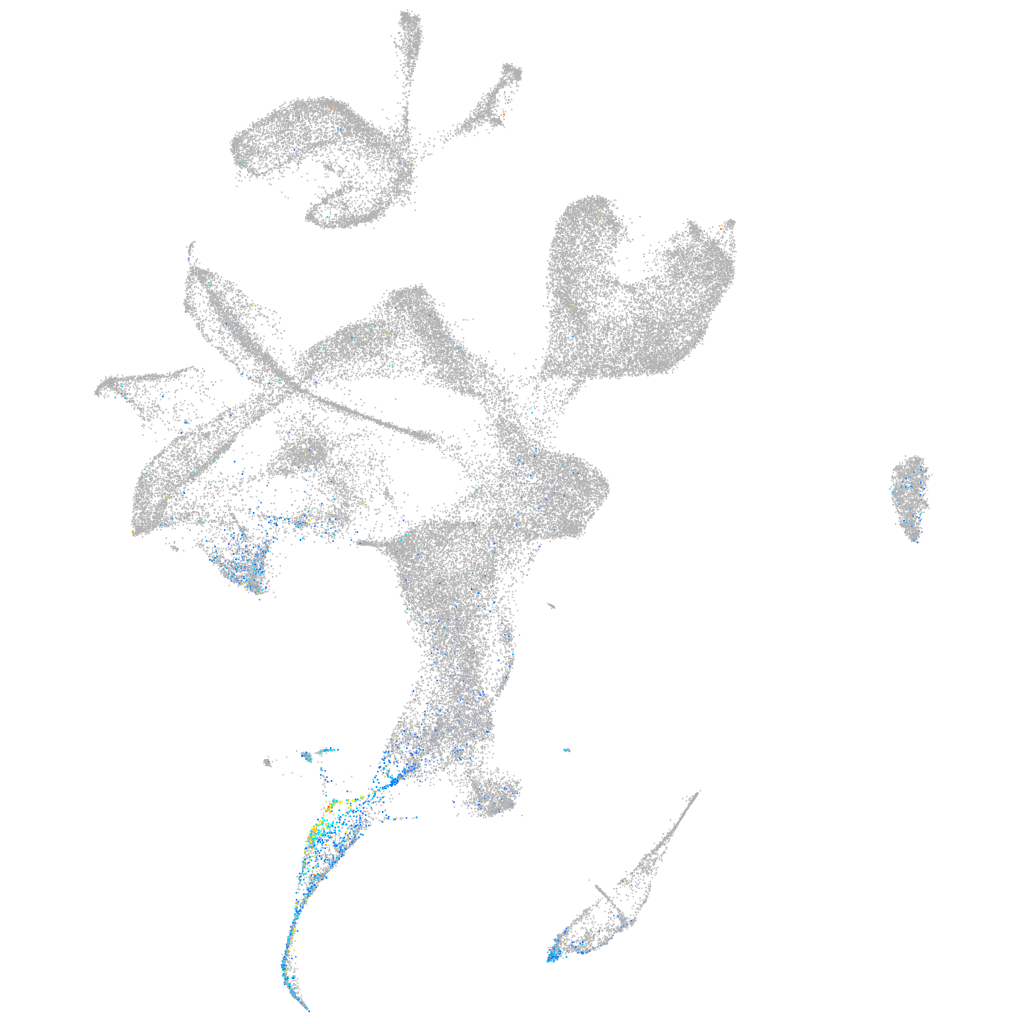
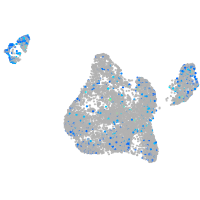
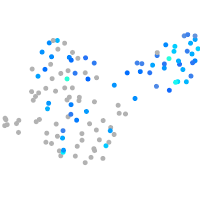
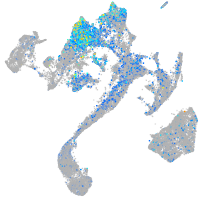
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| pmela | 0.433 | ptmab | -0.220 |
| tyrp1b | 0.431 | si:ch73-1a9.3 | -0.178 |
| dct | 0.427 | hmgn6 | -0.142 |
| tspan36 | 0.425 | tuba1c | -0.132 |
| slc45a2 | 0.419 | gpm6aa | -0.128 |
| rab38 | 0.399 | marcksl1b | -0.127 |
| sytl2b | 0.395 | si:ch211-137a8.4 | -0.124 |
| tspan10 | 0.395 | ptmaa | -0.123 |
| tyrp1a | 0.392 | hmgb1a | -0.112 |
| sparc | 0.391 | tuba1a | -0.105 |
| bace2 | 0.387 | h2afva | -0.104 |
| pmelb | 0.386 | cadm3 | -0.099 |
| gpr143 | 0.378 | rorb | -0.098 |
| gstp1 | 0.377 | hmgn2 | -0.098 |
| oca2 | 0.372 | anp32e | -0.097 |
| col4a5 | 0.370 | fabp7a | -0.096 |
| fstl1b | 0.366 | nova2 | -0.093 |
| cx43 | 0.363 | ckbb | -0.092 |
| rab27a | 0.357 | epb41a | -0.092 |
| cldn19 | 0.355 | si:ch1073-429i10.3.1 | -0.089 |
| col4a6 | 0.353 | foxg1b | -0.089 |
| qdpra | 0.352 | marcksb | -0.087 |
| cracr2ab | 0.347 | hnrnpaba | -0.087 |
| pttg1ipb | 0.347 | snap25b | -0.086 |
| cd63 | 0.343 | neurod4 | -0.085 |
| agtrap | 0.341 | h3f3d | -0.084 |
| comtb | 0.339 | hmgb3a | -0.083 |
| col9a1b | 0.337 | cirbpb | -0.080 |
| slc24a5 | 0.335 | gpm6ab | -0.079 |
| triobpa | 0.335 | stmn1b | -0.079 |
| tmem98 | 0.331 | hmgb1b | -0.077 |
| wnt2 | 0.330 | ndrg4 | -0.077 |
| rnaseka | 0.329 | rtn1a | -0.077 |
| col9a2 | 0.327 | crx | -0.076 |
| zgc:110591 | 0.327 | si:dkey-276j7.1 | -0.075 |